5KMU
 
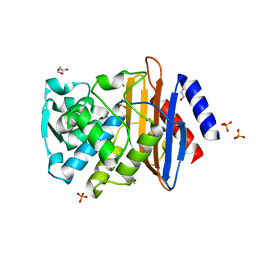 | | CTX-M-9 beta lactamase mutant - T165W | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Latallo, M, Faham, S, Kasson, P.M. | | Deposit date: | 2016-06-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting allosteric mutants that increase activity of a major antibiotic resistance enzyme.
Chem Sci, 8, 2017
|
|
5KMT
 
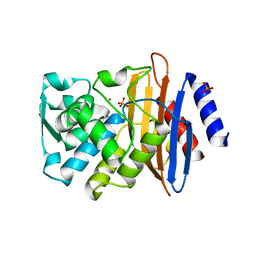 | | CTX-M9 mutant L48A | | Descriptor: | Beta-lactamase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Latallo, M.J, Faham, S, Kasson, P.M. | | Deposit date: | 2016-06-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Predicting allosteric mutants that increase activity of a major antibiotic resistance enzyme.
Chem Sci, 8, 2017
|
|
2MLH
 
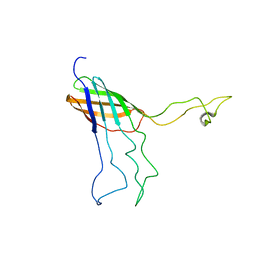 | | NMR Solution Structure of Opa60 from N. Gonorrhoeae in FC-12 Micelles | | Descriptor: | Opacity protein opA60 | | Authors: | Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M, Columbus, L. | | Deposit date: | 2014-02-27 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the neisserial outer membrane protein opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
2MAF
 
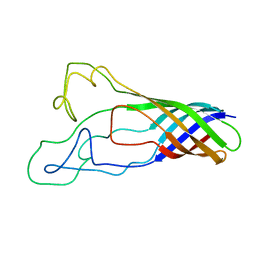 | | Solution structure of opa60 from n. gonorrhoeae | | Descriptor: | Opacity protein opA60 | | Authors: | Columbus, L, Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M. | | Deposit date: | 2013-07-08 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neisserial outer membrane protein Opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
