1GQ4
 
 | |
1GQ5
 
 | |
1I92
 
 | | STRUCTURAL BASIS OF THE NHERF PDZ1-CFTR INTERACTION | | Descriptor: | CHLORIDE ION, NA+/H+ EXCHANGE REGULATORY CO-FACTOR | | Authors: | Karthikeyan, S, Leung, T, Ladias, J.A.A. | | Deposit date: | 2001-03-16 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator.
J.Biol.Chem., 276, 2001
|
|
3B7M
 
 | |
1BIY
 
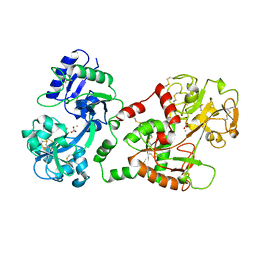 | | STRUCTURE OF DIFERRIC BUFFALO LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Karthikeyan, S, Yadav, S, Singh, T.P. | | Deposit date: | 1998-06-21 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structure of buffalo lactoferrin at 3.3 A resolution at 277 K.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1G9O
 
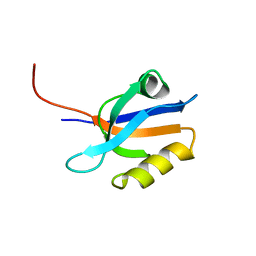 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
1CE2
 
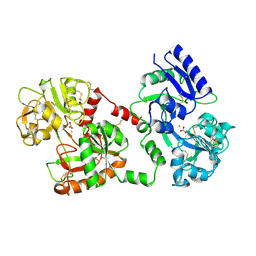 | | STRUCTURE OF DIFERRIC BUFFALO LACTOFERRIN AT 2.5A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (LACTOFERRIN) | | Authors: | Karthikeyan, S, Paramasivam, M, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-03-13 | | Release date: | 1999-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of buffalo lactoferrin at 2.5 A resolution using crystals grown at 303 K shows different orientations of the N and C lobes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1P4M
 
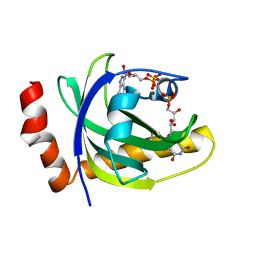 | | CRYSTAL STRUCTURE OF RIBOFLAVIN KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1NB9
 
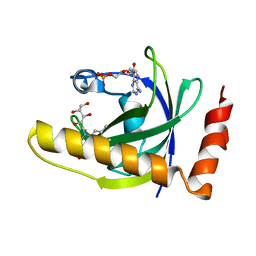 | | Crystal Structure of Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOFLAVIN, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1NB0
 
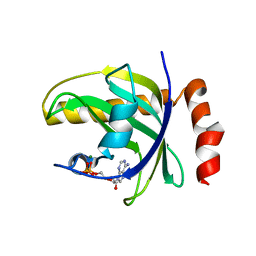 | | Crystal Structure of Human Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein FLJ11149 | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-01 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1Q9S
 
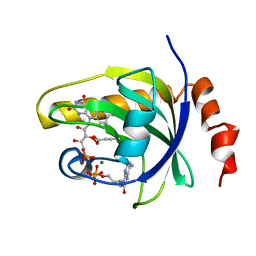 | | Crystal structure of riboflavin kinase with ternary product complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Karthikeyan, S, Zhou, Q, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-08-25 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism.
Biochemistry, 42, 2003
|
|
1RQX
 
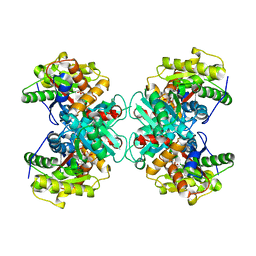 | | Crystal structure of ACC Deaminase complexed with Inhibitor | | Descriptor: | 1-AMINOCYCLOPROPYLPHOSPHONATE, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhao, Z, Kao, C.L, Zhou, Q, Tao, Z, Zhang, H, Liu, H.W. | | Deposit date: | 2003-12-07 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of 1-aminocyclopropane-1-carboxylate deaminase: observation of an aminyl intermediate and identification of Tyr 294 as the active-site nucleophile.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1TZ2
 
 | | Crystal structure of 1-aminocyclopropane-1-carboyxlate deaminase complexed with ACC | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1TZM
 
 | | Crystal structure of ACC deaminase complexed with substrate analog b-chloro-D-alanine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 3-chloro-D-alanine, AMINO-ACRYLATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1TZJ
 
 | | Crystal Structure of 1-aminocyclopropane-1-carboxylate deaminase complexed with d-vinyl glycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, D-VINYLGLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1TYZ
 
 | | Crystal structure of 1-Aminocyclopropane-1-carboyxlate Deaminase from Pseudomonas | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes:Insight into the mechanism of unique pyrodoxial-5'-phosphate dependent cyclopropane ring opening reaction
Biochemistry, 43, 2004
|
|
1TZK
 
 | | Crystal structure of 1-aminocyclopropane-1-carboxylate-deaminase complexed with alpha-keto-butyrate | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 2-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
8H5S
 
 | | Crystal structure of Rv3400 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, CHLORIDE ION, ... | | Authors: | Singh, L, Karthikeyan, S, Thakur, K.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization reveals Rv3400 codes for beta-phosphoglucomutase in Mycobacterium tuberculosis.
Protein Sci., 33, 2024
|
|
8BR4
 
 | | Structure of GAPDH from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, A, Karthikeyan, S. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Stoichiometry of ligand binding and role of C-terminal lysines in Mycobacterium tuberculosis and human GAPDH multifunctionality.
Febs J., 2024
|
|
4RL4
 
 | |
4I14
 
 | | Crystal Structure of Mtb-ribA2 (Rv1415) | | Descriptor: | Riboflavin biosynthesis protein RibBA, SULFATE ION, ZINC ION | | Authors: | Singh, M, Kumar, P, Yadav, S, Gautam, R, Sharma, N, Karthikeyan, S. | | Deposit date: | 2012-11-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure reveals the molecular mechanism of bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II (Rv1415) from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5I2U
 
 | | Crystal structure of a novel Halo-Tolerant Cellulase from Soil Metagenome | | Descriptor: | Cellulase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garg, R, Brahma, V, Srivastava, R, Verma, L, Karthikeyan, S, Sahni, G. | | Deposit date: | 2016-02-09 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome
Sci Rep, 6, 2016
|
|
5HWI
 
 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
5HWK
 
 | | Crystal structure of gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | BENZOIC ACID, Glutathione-specific gamma-glutamylcyclotransferase, PHOSPHATE ION | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
3TZ6
 
 | | Crystal structure of Aspartate semialdehyde dehydrogenase Complexed With inhibitor SMCS (CYS) And Phosphate From Mycobacterium tuberculosis H37Rv | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, GLYCEROL, ... | | Authors: | Vyas, R, Tewari, R, Weiss, M.S, Karthikeyan, S. | | Deposit date: | 2011-09-27 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) from Mycobacterium tuberculosis H37Rv
Acta Crystallogr.,Sect.D, 68, 2012
|
|
