2F3I
 
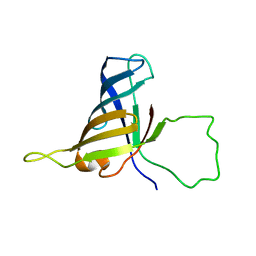 | | Solution Structure of a Subunit of RNA Polymerase II | | Descriptor: | DNA-directed RNA polymerases I, II, and III 17.1 kDa polypeptide | | Authors: | Kang, X, Jin, C. | | Deposit date: | 2005-11-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, biochemical, and dynamic characterizations of the hRPB8 subunit of human RNA polymerases
J.Biol.Chem., 281, 2006
|
|
8QEW
 
 | |
8YK7
 
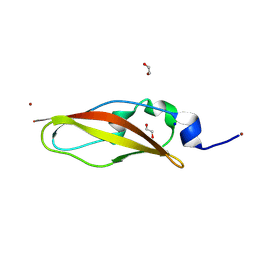 | | Structure of Rib domain from surface adhesin of Limosilactobacillus reuteri | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the Rib domain of the cell-wall-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9AVK
 
 | | Structure of long Rib domain from Limosilactobacillus reuteri | | Descriptor: | SODIUM ION, THIOCYANATE ION, YSIRK signal domain/LPXTG anchor domain surface protein | | Authors: | Xue, Y, Kang, X. | | Deposit date: | 2024-03-04 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structure of the long Rib domain of the LPXTG-anchored surface protein from Limosilactobacillus reuteri.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3EBN
 
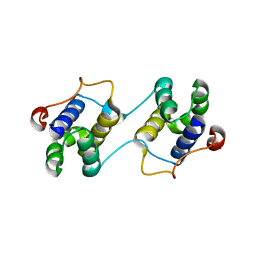 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | Deposit date: | 2008-08-28 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
5HQT
 
 | | Crystal structure of an aspartate/glutamate racemase from Escherichia coli O157 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5HRA
 
 | | Crystal structure of an aspartate/glutamate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
2LIZ
 
 | |
5HRC
 
 | | Crystal structure of an aspartate/glutamate racemase in complex with L-aspartate | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
5HXD
 
 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | Descriptor: | CACODYLATE ION, Protein MpaA, ZINC ION | | Authors: | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
