7D8O
 
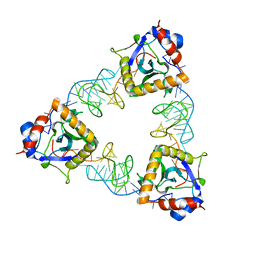 | | Crystal structure of E. coli ToxIN type III toxin-antitoxin complex | | Descriptor: | Antitoxin RNA, Type III toxin-antitoxin system ToxN/AbiQ family toxin | | Authors: | Manikandan, P, Rothweiler, U, Singh, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Identification, functional characterization, assembly and structure of ToxIN type III toxin-antitoxin complex from E. coli.
Nucleic Acids Res., 50, 2022
|
|
1OPP
 
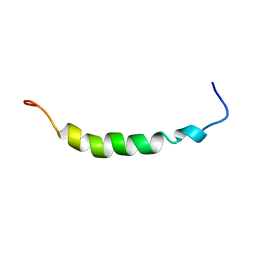 | | PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES | | Descriptor: | APOLIPOPROTEIN C-I | | Authors: | Rozek, A, Buchko, G.W, Kanda, P, Cushley, R.J. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational studies of the N-terminal lipid-associating domain of human apolipoprotein C-I by CD and 1H NMR spectroscopy.
Protein Sci., 6, 1997
|
|
1EZE
 
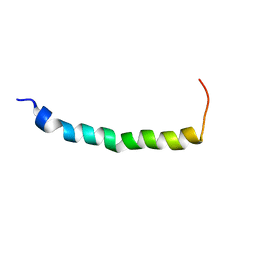 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
6CX6
 
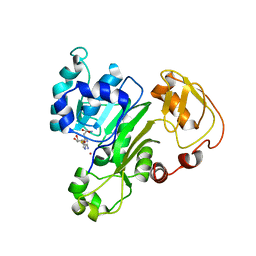 | |
5DFG
 
 | |
5DRH
 
 | | Crystal structure of apo C-As lyase | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, SODIUM ION | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of apo C-As lyase
To Be Published
|
|
4XAE
 
 | | Structure of Feruloyl-CoA 6-hydroxylase (F6H) from Arabidopsis thaliana | | Descriptor: | Feruloyl CoA ortho-hydroxylase 1, SODIUM ION | | Authors: | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Rose, J, Yan, Y. | | Deposit date: | 2014-12-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.769 Å) | | Cite: | Structural Insights into Substrate Specificity of Feruloyl-CoA 6'-Hydroxylase from Arabidopsis thaliana.
Sci Rep, 5, 2015
|
|
6M7G
 
 | | Crystal structure of ArsN, N-acetyltransferase with substrate phosphinothricin from Pseudomonas putida KT2440 | | Descriptor: | PHOSPHINOTHRICIN, Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.657 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
4RSR
 
 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with trivalent phenyl arsencial derivative-Roxarsone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-arsanyl-2-nitrophenol, Arsenic methyltransferase, ... | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4KW7
 
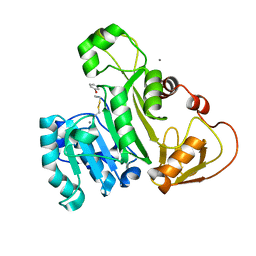 | | The structure of an As(III) S-adenosylmethionine methyltransferase with Phenylarsine oxide(PAO) | | Descriptor: | Arsenic methyltransferase, CALCIUM ION, Phenylarsine oxide | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2013-05-23 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5V0F
 
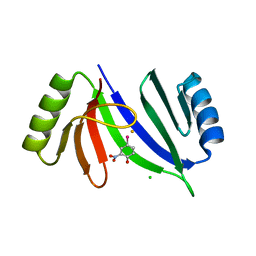 | | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone | | Descriptor: | 4-arsanyl-2-nitrophenol, CHLORIDE ION, FE (III) ION, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2017-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of C-As lyase with mutation K105A and substrate Roxarsone.
To Be Published
|
|
5EVJ
 
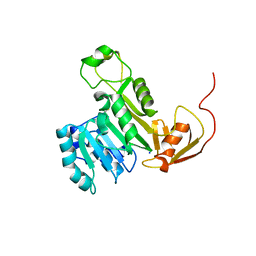 | | X-ray crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii | | Descriptor: | Arsenite methyltransferase, SODIUM ION | | Authors: | Packianathan, C, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii
To Be Published
|
|
6EB0
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, ACETATE ION | | Authors: | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Yan, Y, Rose, J. | | Deposit date: | 2018-08-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
5EG5
 
 | |
6XA0
 
 | | Crystal structure of C-As lyase with mutation K105R with Ni(II) | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase, NICKEL (II) ION | | Authors: | Venkadesh, S, Yoshinaga, M, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The ArsI C-As lyase: Elucidating the catalytic mechanism of degradation of organoarsenicals.
J.Inorg.Biochem., 232, 2022
|
|
6XCK
 
 | | Crystal structure of C-As lyase with mutation K105E | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Venkadesh, S, Yoshinaga, M, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The ArsI C-As lyase: Elucidating the catalytic mechanism of degradation of organoarsenicals.
J.Inorg.Biochem., 232, 2022
|
|
5JTF
 
 | | Crystal structure of ArsN N-acetyltransferase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
5JX7
 
 | | Cysteine mutant (C224A) structure of As (III) S-adenosyl methyltransferase | | Descriptor: | Arsenic methyltransferase, CALCIUM ION | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine mutant (C224A) structure of As (III) S-adenosyl methyltransferase
To be Published
|
|
5HCW
 
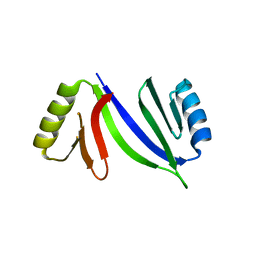 | |
6B1B
 
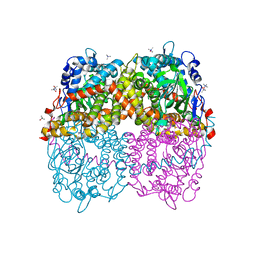 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI MUTANT XS6 (APO Enzyme) | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, trimethylamine oxide | | Authors: | Zhou, D, Kandavelu, P, Wang, B.C, Yan, Y, Rose, J.P. | | Deposit date: | 2017-09-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
5C68
 
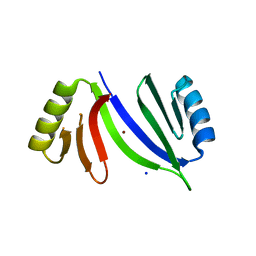 | |
5C4P
 
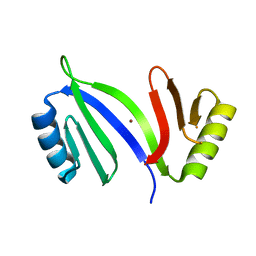 | |
5C6X
 
 | | Crystal structure of C-As lyase with Co(II) | | Descriptor: | COBALT (II) ION, GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-23 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of C-As lyase with Co(II)
To Be Published
|
|
5CB9
 
 | | Crystal structure of C-As lyase with mercaptoethonal | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Glyoxalase/bleomycin resistance protein/dioxygenase, ... | | Authors: | Venkadesh, S, Yoshinaga, M, Sankaran, B, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2015-06-30 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of C-As lyase with mercaptoethonal
To Be Published
|
|
6J05
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | ARSENIC, SODIUM ION, Transcriptional regulator ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
