1IRM
 
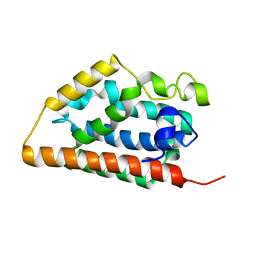 | | Crystal structure of apo heme oxygenase-1 | | Descriptor: | apo heme oxygenase-1 | | Authors: | Sugishima, M, Sakamoto, H, Kakuta, Y, Omata, Y, Hayashi, S, Noguchi, M, Fukuyama, K. | | Deposit date: | 2001-10-09 | | Release date: | 2002-07-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of rat apo-heme oxygenase-1 (HO-1): mechanism of heme binding in HO-1 inferred from structural comparison of the apo and heme complex forms
Biochemistry, 41, 2002
|
|
5Z8B
 
 | |
5X2B
 
 | |
8KDA
 
 | | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, MAGNESIUM ION, RNA-free ribonuclease P | | Authors: | Teramoto, T, Adachi, N, Yokogawa, T, Koyasu, T, Mayanagi, K, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure of Hydrogenobacter thermophilus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
8KD9
 
 | | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs | | Descriptor: | Aquifex aeolicus pre-tRNAVal, RNA-free ribonuclease P | | Authors: | Teramoto, T, Koyasu, T, Mayanagi, K, Yokogawa, T, Adachi, N, Nakamura, T, Senda, T, Kakuta, Y. | | Deposit date: | 2023-08-09 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure of Aquifex aeolicus minimal protein-only RNase P (HARP) in complex with pre-tRNAs
To Be Published
|
|
8XF0
 
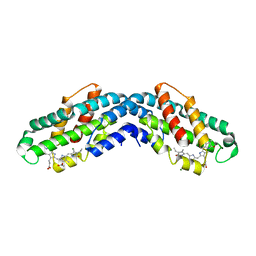 | | Crystal structure of the dissociated C-phycocyanin alpha-chain from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, MAGNESIUM ION, PHYCOCYANOBILIN | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
8XF2
 
 | | Crystal structure of the reassembled C-phycocyanin hexamer from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHOSPHATE ION, ... | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
8XF1
 
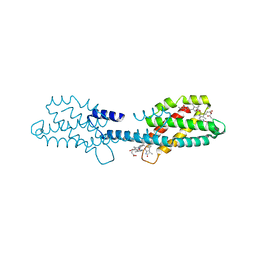 | | Crystal structure of the dissociated C-phycocyanin beta-chain from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
8XF3
 
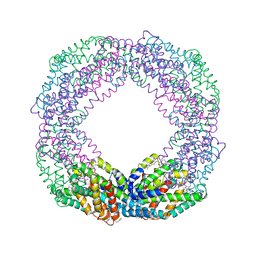 | | Crystal structure of the reassembled C-phycocyanin octamer from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHOSPHATE ION, ... | | Authors: | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
8Y9P
 
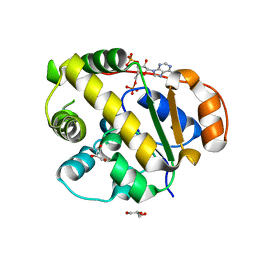 | |
1IQZ
 
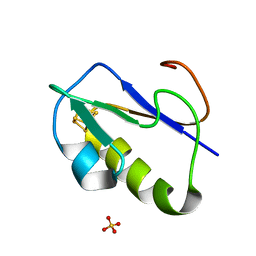 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM I) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
1IR0
 
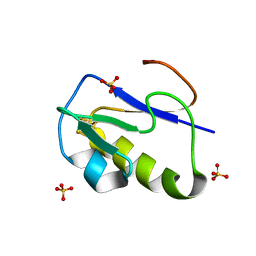 | | OXIDIZED [4Fe-4S] FERREDOXIN FROM BACILLUS THERMOPROTEOLYTICUS (FORM II) | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Fukuyama, K, Okada, T, Kakuta, Y, Takahashi, Y. | | Deposit date: | 2001-08-30 | | Release date: | 2002-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: systematic distortion of [4Fe-4S] cluster in the protein.
J.Mol.Biol., 315, 2002
|
|
3VKK
 
 | | Crystal Structure Of The Covalent Intermediate Of Human Cytosolic Beta-Glucosidase-mannose complex | | Descriptor: | CHLORIDE ION, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Okino, N, Ito, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition mechanism of human cytosolic beta-glucosidase by monnoside
To be Published
|
|
1CJM
 
 | | HUMAN SULT1A3 WITH SULFATE BOUND | | Descriptor: | PROTEIN (ARYL SULFOTRANSFERASE), SULFATE ION | | Authors: | Bidwell, L.M, Mcmanus, M.E, Gaedigk, A, Kakuta, Y, Negishi, M, Pedersen, L, Martin, J.L. | | Deposit date: | 1999-04-18 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human catecholamine sulfotransferase.
J.Mol.Biol., 293, 1999
|
|
1IR6
 
 | | Crystal structure of exonuclease RecJ bound to manganese | | Descriptor: | MANGANESE (II) ION, exonuclease RecJ | | Authors: | Yamagata, A, Kakuta, Y, Masui, R, Fukuyama, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-09-11 | | Release date: | 2002-05-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6K3N
 
 | |
1IYZ
 
 | |
1IZ0
 
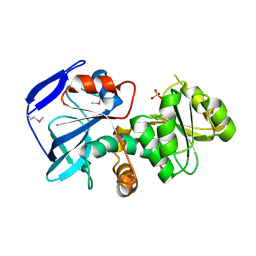 | |
1J1F
 
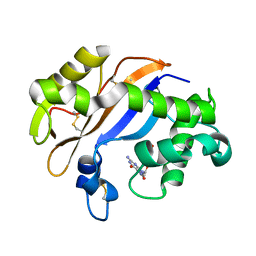 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1G
 
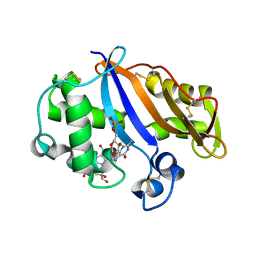 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
3VU2
 
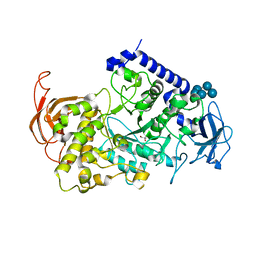 | | Structure of the Starch Branching Enzyme I (BEI) complexed with maltopentaose from Oryza sativa L | | Descriptor: | 1,4-alpha-glucan-branching enzyme, chloroplastic/amyloplastic, GLYCEROL, ... | | Authors: | Chaen, K, Kakuta, Y, Kimura, M. | | Deposit date: | 2012-06-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of the rice branching enzyme I (BEI) in complex with maltopentaose.
Biochem.Biophys.Res.Commun., 424, 2012
|
|
2ZOX
 
 | | Crystal Structure of the Covalent Intermediate of Human Cytosolic beta-Glucosidase | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Baba, Y, Okino, N, Kimura, M, Ito, M, Kakuta, Y. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase
Biochem.Biophys.Res.Commun., 374, 2008
|
|
7EOV
 
 | | Crystal structure of mouse cytosolic sulfotransferase mSULT2A8 in complex with PAP and cholic acid | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CHOLIC ACID, cytosolic sulfotransferase SULT2A8 | | Authors: | Teramoto, T, Nishio, T, Kakuta, Y. | | Deposit date: | 2021-04-22 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of mouse SULT2A8 reveals the mechanism of 7 alpha-hydroxyl, bile acid sulfation.
Biochem.Biophys.Res.Commun., 562, 2021
|
|
1IYB
 
 | |
1UAX
 
 | |
