1Y1Q
 
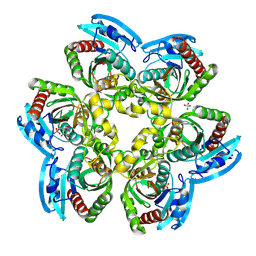 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Uridine-5p-monophosphate and Sulfate Ion at 2.35A Resolution | | Descriptor: | SULFATE ION, URIDINE-5'-MONOPHOSPHATE, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
1Y1R
 
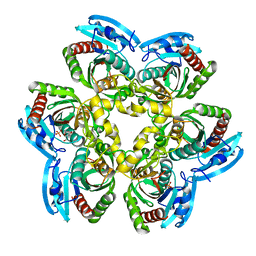 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Inhibitor and Phosphate Ion at 2.11A Resolution | | Descriptor: | 2,2'-Anhydro-(1-beta-D-ribofuranosyl)uracil, PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M.V, Gabdoulkhakov, A.G, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Complex with Inhibitor and Phosphate.
To be Published
|
|
1Y1S
 
 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with Uracil and Sulfate Ion at 2.55A Resolution | | Descriptor: | SULFATE ION, URACIL, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
1Y1T
 
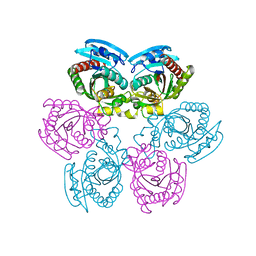 | | Crystal Structure of the Uridine Phosphorylase from Salmonella Typhimurium at 1.77A Resolution | | Descriptor: | GLYCEROL, SULFATE ION, Uridine phosphorylase | | Authors: | Gabdoulkhakov, A.G, Dontsova, M.V, Kachalova, G.S, Betzel, C, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2004-11-19 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structures of Salmonella Typhimurium Uridine Phosphorylase in Native and Three Complexes Forms - with Uridine, Uracil and Sulfate.
To be Published
|
|
7POH
 
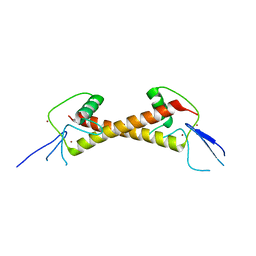 | | Crystal structure of ZAD-domain of Serendipity-d protein from D.melanogaster | | Descriptor: | Serendipity locus protein delta, ZINC ION | | Authors: | Boyko, K.M, Kachalova, G.S, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
6FP5
 
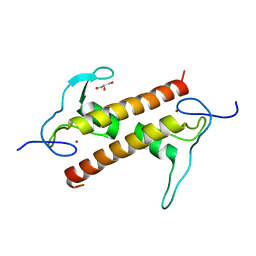 | | Crystal structure of ZAD-domain of CG2712 protein from D.melanogaster | | Descriptor: | CG2712, GLYCEROL, ZINC ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bonchuk, A.N, Kachalova, G.S, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of diversity and homodimerization specificity of zinc-finger-associated domains in Drosophila.
Nucleic Acids Res., 49, 2021
|
|
3K7T
 
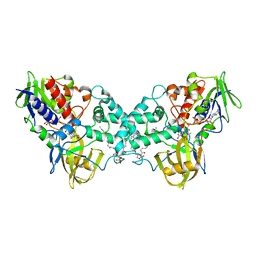 | | Crystal structure of apo-form 6-hydroxy-L-nicotine oxidase, crystal form P3121 | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
2O0B
 
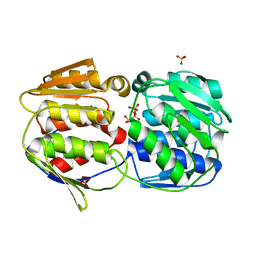 | |
3K7Q
 
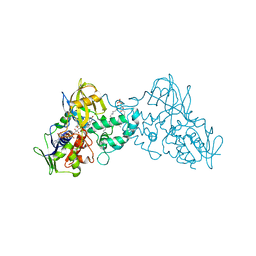 | | Crystal structure of substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
2NNY
 
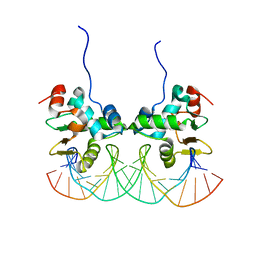 | | Crystal structure of the Ets1 dimer DNA complex. | | Descriptor: | 5'-D(*A*CP*TP*CP*CP*AP*GP*GP*AP*AP*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*TP*CP*T)-3', 5'-D(*T*AP*GP*AP*CP*AP*GP*GP*AP*AP*GP*CP*AP*CP*TP*TP*CP*CP*TP*GP*GP*AP*G)-3', C-ets-1 protein | | Authors: | Lamber, E.P, Kachalova, G.S, Wilmanns, M. | | Deposit date: | 2006-10-24 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization.
Embo J., 27, 2008
|
|
2O2J
 
 | |
2O2E
 
 | |
