7TJ1
 
 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-14 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
7TWC
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, DUF1479 domain-containing protein, GLYCEROL, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to CAPS
To Be Published
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | 分子名称: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | 著者 | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2013-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
4IQI
 
 | | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae O1 biovar El Tor complexed with cytosine | | 分子名称: | 6-AMINOPYRIMIDIN-2(1H)-ONE, CHLORIDE ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | 著者 | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-11 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae O1 biovar El Tor complexed with cytosine
To be Published
|
|
5HPI
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain-3-Hydroxy Benzoic Acid complex from Acinetobacter | | 分子名称: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, SULFATE ION, ... | | 著者 | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.963 Å) | | 主引用文献 | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
5F1P
 
 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | 分子名称: | PtmO8 | | 著者 | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-11-30 | | 公開日 | 2015-12-30 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
5HPF
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain from Acinetobacter | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
4IQE
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor | | 分子名称: | Carboxyvinyl-carboxyphosphonate phosphorylmutase | | 著者 | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-11 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
4IQD
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis | | 分子名称: | Carboxyvinyl-carboxyphosphonate phosphorylmutase, PYRUVIC ACID | | 著者 | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-11 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis
To be Published
|
|
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | 分子名称: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | 著者 | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-12 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
7TWE
 
 | | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DUF1479 domain-containing protein, ... | | 著者 | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-02-07 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal Structure of the Putative Oxidoreductase of DUF1479-containing Protein Family YPO2976 from Yersinia pestis Bound to 2-oxo-glutaric acid
To Be Published
|
|
3KWO
 
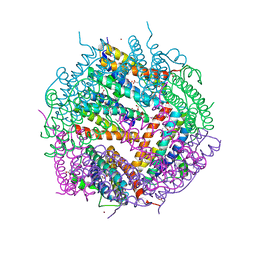 | | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni | | 分子名称: | 1,4-BUTANEDIOL, ACETIC ACID, GLYCEROL, ... | | 著者 | Kim, Y, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-12-01 | | 公開日 | 2010-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.985 Å) | | 主引用文献 | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni
To be Published
|
|
5VVH
 
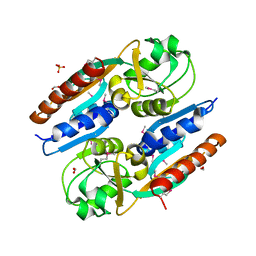 | | Crystal Structure of the Effector Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens | | 分子名称: | FORMIC ACID, Octopine catabolism/uptake operon regulatory protein OccR, SULFATE ION | | 著者 | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-21 | | 最終更新日 | 2018-09-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
2QQY
 
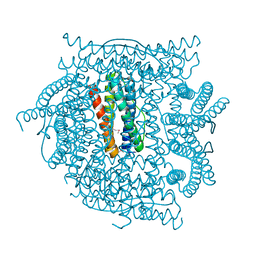 | | Crystal structure of ferritin like, diiron-carboxylate proteins from Bacillus anthracis str. Ames | | 分子名称: | Sigma B operon | | 著者 | Kim, Y, Joachimiak, G, Wu, R, Patterson, S, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-07-27 | | 公開日 | 2007-08-14 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Ferritin like, Diiron-carboxylate Proteins from Bacillus anthracis str. Ames.
To be Published
|
|
5BMZ
 
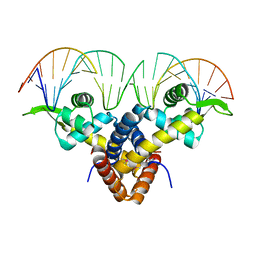 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA. | | 分子名称: | DNA (5'-D(P*GP*AP*AP*TP*AP*TP*CP*AP*GP*TP*TP*AP*AP*AP*CP*TP*GP*AP*TP*AP*TP*TP*C)-3'), HcaR protein | | 著者 | Kim, Y, Joachimiak, G, Biglow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA.
To Be Published
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | 分子名称: | GLYCEROL, Putative aminotransferase | | 著者 | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-19 | | 公開日 | 2015-06-03 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
4IQF
 
 | | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, Methionyl-tRNA formyltransferase, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-11 | | 公開日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (2.378 Å) | | 主引用文献 | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis
To be Published
|
|
4J01
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 29mer DNA target | | 分子名称: | DNA (29-MER), SULFATE ION, Transcription Factor HetR | | 著者 | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.246 Å) | | 主引用文献 | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J2Q
 
 | |
5VUG
 
 | | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase Domain of Uncharacterized Protein Rv2277c from Mycobacterium tuberculosis | | 分子名称: | CALCIUM ION, GLYCEROL, Uncharacterized protein Rv2277c | | 著者 | Kim, Y, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-14 | | 最終更新日 | 2017-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase Domain of Uncharacterized Protein Rv2277c from Mycobacterium tuberculosis
To Be Published
|
|
4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | 著者 | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5VVI
 
 | | Crystal Structure of the Ligand Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens in the Complex with Octopine | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
4IXH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum | | 分子名称: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-25 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Optimization of Benzoxazole-Based Inhibitors of Cryptosporidium parvum Inosine 5'-Monophosphate Dehydrogenase.
J.Med.Chem., 56, 2013
|
|
4J00
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | 分子名称: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | 著者 | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2013-01-30 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.004 Å) | | 主引用文献 | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1WC8
 
 | | The crystal structure of mouse bet3p | | 分子名称: | MYRISTIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT3 | | 著者 | Kim, Y.-G, Sacher, M, Oh, B.-H. | | 登録日 | 2004-11-10 | | 公開日 | 2004-12-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Bet3 Reveals a Novel Mechanism for Golgi Localization of Tethering Factor Trapp
Nat.Struct.Mol.Biol., 12, 2005
|
|
