1VLH
 
 | |
1VM8
 
 | |
1VJ0
 
 | |
1VLV
 
 | |
1VM6
 
 | |
1VPH
 
 | |
2GPJ
 
 | |
1VKD
 
 | |
1VME
 
 | |
1VPA
 
 | |
1VMJ
 
 | |
2F6R
 
 | |
1O5L
 
 | |
3UWS
 
 | |
4QDG
 
 | |
2ISB
 
 | |
4Q98
 
 | |
4QB7
 
 | |
2GB3
 
 | |
4QJR
 
 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to its hormone pip3 at 2.4 a resolution | | Descriptor: | (2S)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dihexadecanoate, ACETATE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QK4
 
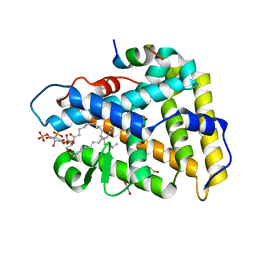 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to pip2 at 2.8 a resolution | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-05 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1VJT
 
 | |
1VKK
 
 | |
1VKU
 
 | |
2H56
 
 | |
