6LXD
 
 | | Pri-miRNA bound DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, RNA (102-mer), Ribonuclease 3, ... | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
6LXE
 
 | | DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3, ZINC ION | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
5H1L
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 7-nt U4 snRNA fragment | | Descriptor: | GLYCEROL, Gem-associated protein 5, U4 snRNA (5'-R(*AP*UP*UP*UP*UP*UP*G)-3') | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1J
 
 | | Crystal structure of WD40 repeat domains of Gemin5 | | Descriptor: | Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5H1M
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with M7G | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Gem-associated protein 5 | | Authors: | Jin, W, Wang, Y, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
1TV0
 
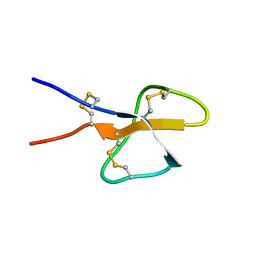 | | Solution structure of cryptdin-4, the most potent alpha-defensin from mouse Paneth cells | | Descriptor: | Cryptdin-4 | | Authors: | Jing, W, Hunter, H.N, Tanabe, H, Ouellette, A.J, Vogel, H.J. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Biochemistry, 43, 2004
|
|
2RTU
 
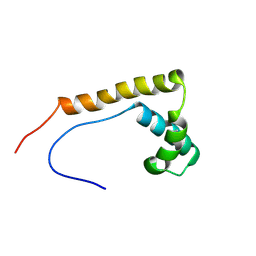 | | Solution structure of oxidized human HMGB1 A box | | Descriptor: | High mobility group protein B1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA.
Biochem.Biophys.Res.Commun., 441, 2013
|
|
2RUQ
 
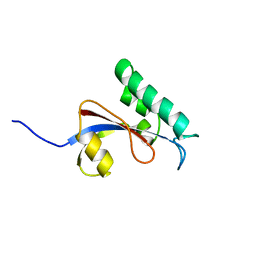 | | solution structure of human Pin1 PPIase mutant C113A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
2RUR
 
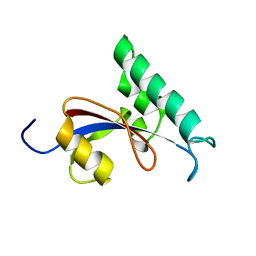 | | Solution structure of Human Pin1 PPIase C113S mutant | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric Breakage of the Hydrogen Bond within the Dual-Histidine Motif in the Active Site of Human Pin1 PPIase
Biochemistry, 54, 2015
|
|
1LIR
 
 | | LQ2 FROM LEIURUS QUINQUESTRIATUS, NMR, 22 STRUCTURES | | Descriptor: | LQ2 | | Authors: | Renisio, J.G, Lu, Z, Blanc, E, Jin, W, Lewis, J.H, Bornet, O, Darbon, H. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of potassium channel-inhibiting scorpion toxin Lq2.
Proteins, 34, 1999
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
4QYY
 
 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
2DOO
 
 | | The structure of IMP-1 complexed with the detecting reagent (DansylC4SH) by a fluorescent probe | | Descriptor: | BETA-LACTAMASE IMP-1, N-[4-({[5-(DIMETHYLAMINO)-1-NAPHTHYL]SULFONYL}AMINO)BUTYL]-3-SULFANYLPROPANAMIDE, ZINC ION | | Authors: | Kurosaki, H, Yamaguchi, Y, Yasuzawa, H, Jin, W, Yamagata, Y, Arakawa, Y. | | Deposit date: | 2006-05-01 | | Release date: | 2006-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups
Chemmedchem, 1, 2006
|
|
5H1K
 
 | | Crystal structure of WD40 repeat domains of Gemin5 in complex with 13-nt U4 snRNA fragment | | Descriptor: | Gem-associated protein 5, U4 snRNA (5'-R(*GP*CP*AP*AP*UP*UP*UP*UP*UP*GP*AP*CP*A)-3') | | Authors: | Wang, Y, Jin, W, Liu, C.P, Yang, N, Jin, M, Cong, Y, Wang, M, Xu, R.M. | | Deposit date: | 2016-10-10 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5
Genes Dev., 30, 2016
|
|
5GWA
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2016-09-09 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
5GS8
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2016-08-14 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
5X5G
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
2JNR
 
 | | Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide | | Descriptor: | ENV polyprotein, VIR165 | | Authors: | Munch, J, Standker, L, Adermann, K, Schulz, A, Pohlmann, S, Chaipan, C, Biet, T, Peters, T, Meyer, B, Wilhelm, D, Lu, H, Jing, W, Jiang, S, Forssmann, W, Kirchhoff, F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Cell(Cambridge,Mass.), 129, 2007
|
|
