2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | 分子名称: | Protein tyrosine phosphatase type IVA 3 | | 著者 | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | 登録日 | 2013-07-29 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
9KOK
 
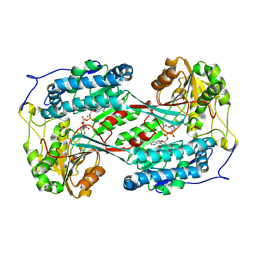 | | Crystal structure of ExaC, an NAD+-dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa | | 分子名称: | GLYCEROL, MAGNESIUM ION, NAD+ dependent aldehyde dehydrogenase ExaC, ... | | 著者 | Lee, J.Y, Ko, J.H, Jeong, K.H, Son, S.B. | | 登録日 | 2024-11-20 | | 公開日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of ExaC, an NAD + -dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 742, 2025
|
|
3U0R
 
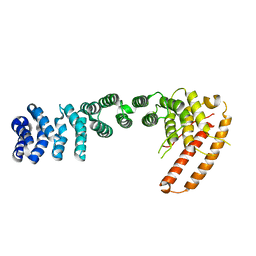 | | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules | | 分子名称: | Apoptosis inhibitor 5 | | 著者 | Han, B.G, Kim, K.H, Jeong, K.C, Cho, J.W, Noh, K.H, Kim, T.W, Yoon, H.J, Suh, S.W, Lee, S.H, Lee, B.I. | | 登録日 | 2011-09-29 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Helical repeat structure of apoptosis inhibitor 5 reveals protein-protein interaction modules.
J.Biol.Chem., 287, 2012
|
|
9J8E
 
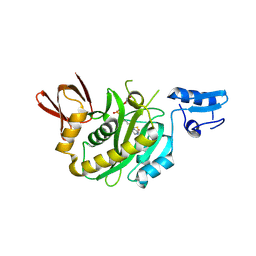 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | 分子名称: | BIOTINYL-5-AMP, Bifunctional ligase/repressor BirA | | 著者 | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | 登録日 | 2024-08-21 | | 公開日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9J8F
 
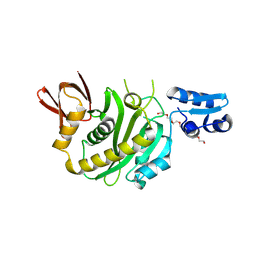 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bifunctional ligase/repressor BirA, PENTAETHYLENE GLYCOL | | 著者 | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | 登録日 | 2024-08-21 | | 公開日 | 2024-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9KOI
 
 | | Crystal structure of ExaC, an NAD+-dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa | | 分子名称: | NAD+ dependent aldehyde dehydrogenase ExaC | | 著者 | Lee, J.Y, Ko, J.H, Jeong, K.H, Son, S.B. | | 登録日 | 2024-11-20 | | 公開日 | 2024-12-11 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of ExaC, an NAD + -dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 742, 2025
|
|
7CV2
 
 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | 分子名称: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | 著者 | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | 登録日 | 2020-08-25 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
8KE8
 
 | | Crystal structure of TetR-type transcriptional factor NalC from P. aeruginosa | | 分子名称: | NalC | | 著者 | Lee, J.Y, Jeong, K.H, Ko, J.H, Son, S.B. | | 登録日 | 2023-08-11 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural insights into the transcriptional regulator NalC, a key component of the MexAB-OprM efflux pump system, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 679, 2023
|
|
2N50
 
 | | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis | | 分子名称: | Acyl carrier protein | | 著者 | Park, Y, Jung, M, Song, H, Jeong, K, Kim, Y. | | 登録日 | 2015-07-02 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis.
J. Biol. Chem., 291, 2016
|
|
7CV0
 
 | | Crystal structure of B. halodurans NiaR in apo form | | 分子名称: | Transcriptional regulator NiaR, ZINC ION | | 著者 | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | 登録日 | 2020-08-25 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
2LXK
 
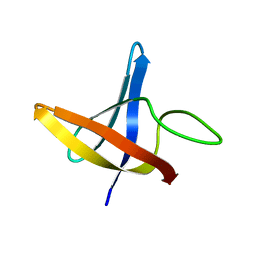 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp | | 分子名称: | Cold shock-like protein CspLA | | 著者 | Lee, J, Jeong, K, Kim, Y. | | 登録日 | 2012-08-27 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
2LXJ
 
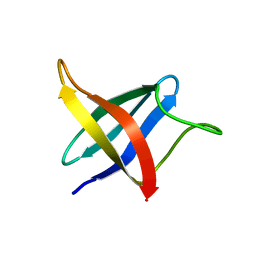 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp with dT7 | | 分子名称: | Cold shock-like protein CspLA | | 著者 | Lee, J, Jeong, K, Kim, Y. | | 登録日 | 2012-08-27 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
7WB3
 
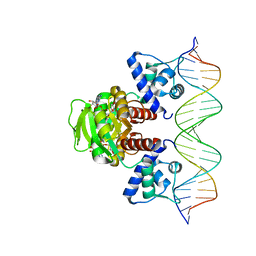 | | Crystal structure of T. maritima Rex in ternary complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*TP*GP*AP*GP*AP*AP*AP*TP*TP*TP*AP*TP*CP*AP*CP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*TP*GP*AP*TP*AP*AP*AP*TP*TP*TP*CP*TP*CP*AP*AP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Lee, J.Y, Jeong, K.H, Lee, H.J, Park, Y.W. | | 登録日 | 2021-12-15 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Structural Basis of Redox-Sensing Transcriptional Repressor Rex with Cofactor NAD + and Operator DNA.
Int J Mol Sci, 23, 2022
|
|
2MO0
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp | | 分子名称: | Cold-shock DNA-binding domain protein | | 著者 | Jin, B, Jeong, K.W, Kim, Y. | | 登録日 | 2014-04-17 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
2MO1
 
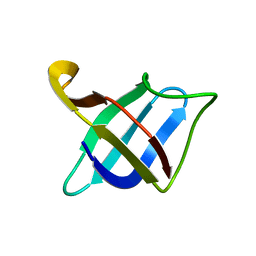 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, TaCsp with dT7 | | 分子名称: | Cold-shock DNA-binding domain protein | | 著者 | Jin, B, Jeong, K.W, Kim, Y. | | 登録日 | 2014-04-17 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and flexibility of the thermophilic cold-shock protein of Thermus aquaticus.
Biochem.Biophys.Res.Commun., 451, 2014
|
|
7D1I
 
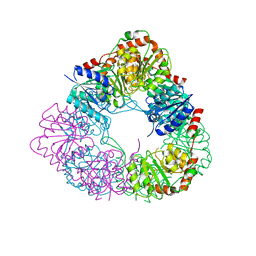 | | Crystal structure of acinetobacter baumannii MurG | | 分子名称: | UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | | 著者 | Park, H.H, Jeong, k.H. | | 登録日 | 2020-09-14 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.487 Å) | | 主引用文献 | Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG
Iucrj, 8, 2021
|
|
7D27
 
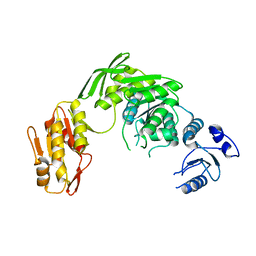 | |
