1SUO
 
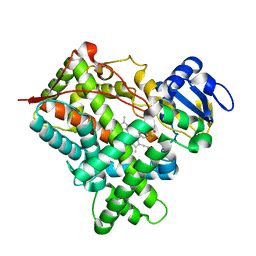 | | Structure of mammalian cytochrome P450 2B4 with bound 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, White, M.A, He, Y.A, Johnson, E.F, Stout, C.D, Halpert, J.R. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 {angstrom} resolution: Insight into the range of P450 conformations and coordination of redox partner binding.
J.Biol.Chem., 279, 2004
|
|
1E6C
 
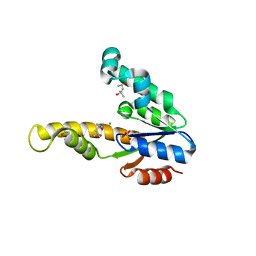 | | K15M MUTANT OF SHIKIMATE KINASE FROM ERWINIA CHRYSANTHEMI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Maclean, J, Krell, T, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2000-08-10 | | Release date: | 2001-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and X-Ray Crystallographic Studies on Shikimate Kinase: The Important Structural Role of the P-Loop Lysine
Protein Sci., 10, 2001
|
|
3ZF2
 
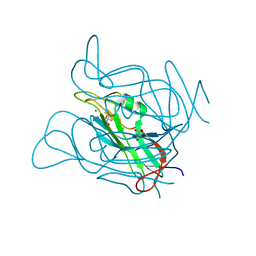 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3QCS
 
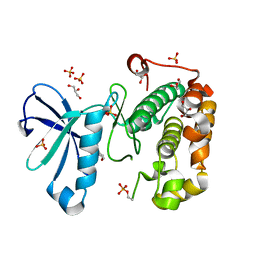 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-[2-Amino-6-(4-morpholinyl)-4-pyrimidinyl]-1H-indazol-3-amine | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 6-[2-amino-6-(morpholin-4-yl)pyrimidin-4-yl]-2H-indazol-3-amine, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QD0
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with (2R,5S)-1-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-N-phenyl-3-piperidinecarboxamide | | Descriptor: | (3S,6R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-6-methyl-N-phenylpiperidine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3ZF6
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81A D110C S168C mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZF1
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81N mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZF5
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y84F mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3S24
 
 | | Crystal structure of human mRNA guanylyltransferase | | Descriptor: | SULFATE ION, mRNA-capping enzyme | | Authors: | Das, K, Chu, C, Thyminski, J.R, Bauman, J.D, Guan, R, Qiu, W, Montelione, G.T, Arnold, E, Shatkin, A.J. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.0137 Å) | | Cite: | Structure of the guanylyltransferase domain of human mRNA capping enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ZF4
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZF3
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y84I mutant). | | Descriptor: | DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZEZ
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism.(Staphylococcus bacteriophage 80alpha dUTPase with dUPNHPP). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
1MBL
 
 | |
1PA0
 
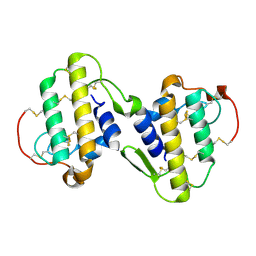 | | CRYSTAL STRUCTURE OF BNSP-7, A LYS49-PHOSPHOLIPASE A2 | | Descriptor: | Myotoxic phospholipase A2-like | | Authors: | Magro, A.J, Soares, A.M, Giglio, J.R, Fontes, M.R. | | Deposit date: | 2003-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of BnSP-7 and BnSP-6, two Lys49-phospholipases A(2): quaternary structure and inhibition mechanism insights.
Biochem.Biophys.Res.Commun., 311, 2003
|
|
2ORE
 
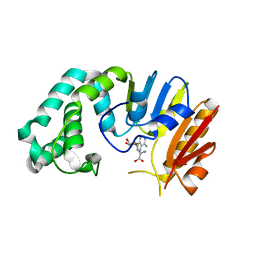 | |
4EQF
 
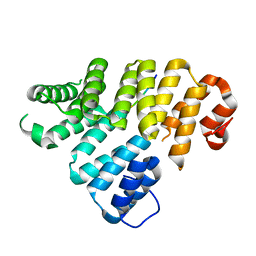 | | Trip8b-1a#206-567 interacting with the carboxy-terminal seven residues of HCN2 | | Descriptor: | PEX5-related protein, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Bankston, J.R, Camp, S.S, Dimaio, F, Lewis, A.S, Chetkovich, D.M, Zagotta, W.N. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EQS
 
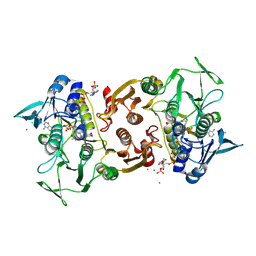 | | Crystal structure of the Y419F mutant of Staphylococcus aureus CoADR | | Descriptor: | CHLORIDE ION, COENZYME A, Coenzyme A disulfide reductase, ... | | Authors: | Wallace, B.D, Edwards, J.S, Wallen, J.R, Claiborne, A, Redinbo, M.R. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Turnover-Dependent Covalent Inactivation of Staphylococcus aureus Coenzyme A-Disulfide Reductase by Coenzyme A-Mimetics: Mechanistic and Structural Insights.
Biochemistry, 51, 2012
|
|
4IYN
 
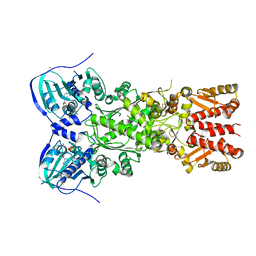 | | Structure of mitochondrial Hsp90 (TRAP1) with ADP-ALF4- | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Partridge, J.R, Lavery, L.A, Agard, D.A. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism.
Mol.Cell, 53, 2014
|
|
1Z6U
 
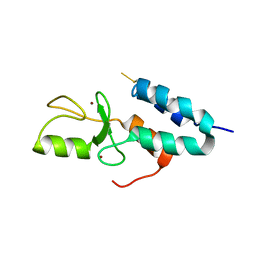 | | Np95-like ring finger protein isoform b [Homo sapiens] | | Descriptor: | Np95-like ring finger protein isoform b, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Crystal Structure of the Human Ubiquitin Liagse NIRF
To be Published
|
|
4ERU
 
 | | Crystal Structure of Putative Cytoplasmic Protein, YciF Bacterial Stress Response Protein from Salmonella enterica | | Descriptor: | D-MALATE, MAGNESIUM ION, YciF Bacterial Stress Response Protein | | Authors: | Kim, Y, Wu, R, Jedrzejczak, R, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-20 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Putative Cytoplasmic Protein, YciF Bacterial Stress Response Protein from Salmonella enterica
To be Published
|
|
3KCI
 
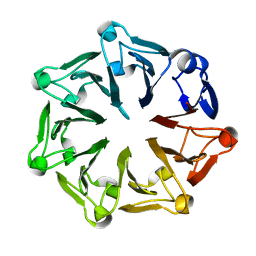 | | The third RLD domain of HERC2 | | Descriptor: | Probable E3 ubiquitin-protein ligase HERC2 | | Authors: | Walker, J.R, Qiu, L, Vesterberg, A, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Third RLD Domain of Herc2
To be Published
|
|
4EVX
 
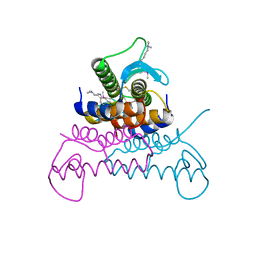 | | Crystal structure of putative phage endolysin from S. enterica | | Descriptor: | Putative phage endolysin | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative phage endolysin from S. enterica
To be Published
|
|
8DQ0
 
 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, RhlR protein | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
1ZB9
 
 | | Crystal structure of Xylella fastidiosa organic peroxide resistance protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, organic hydroperoxide resistance protein | | Authors: | Oliveira, M.A, Guimaraes, B.G, Cussiol, J.R, Medrano, F.J, Vidigal, S.A, Gozzo, F.C, Netto, L.E. | | Deposit date: | 2005-04-07 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Enzyme-Substrate Interaction and Characterization of Enzymatic Intermediates of Organic Hydroperoxide Resistance Protein from Xylella fastidiosa.
J.Mol.Biol., 359, 2006
|
|
3BJZ
 
 | | Crystal structure of Pseudomonas aeruginosa phosphoheptose isomerase | | Descriptor: | CHLORIDE ION, Phosphoheptose isomerase, SULFATE ION | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants.
J.Biol.Chem., 283, 2008
|
|
