3FK9
 
 | | Crystal structure of mMutator MutT protein from Bacillus halodurans | | Descriptor: | Mutator MutT protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mMutator MutT protein from Bacillus halodurans
To be Published
|
|
3FBT
 
 | | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum | | Descriptor: | SULFATE ION, chorismate mutase and shikimate 5-dehydrogenase fusion protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a chorismate mutase/shikimate 5-dehydrogenase fusion protein from Clostridium acetobutylicum
To be Published
|
|
3FDW
 
 | | Crystal structure of a C2 domain from human synaptotagmin-like protein 4 | | Descriptor: | Synaptotagmin-like protein 4 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a C2 domain from human synaptotagmin-like protein 4
To be Published
|
|
1P54
 
 | |
1P8K
 
 | |
1OXW
 
 | | The Crystal Structure of SeMet Patatin | | Descriptor: | Patatin | | Authors: | Rydel, T.J, Williams, J.M, Krieger, E, Moshiri, F, Stallings, W.C, Brown, S.M, Pershing, J.C, Purcell, J.P, Alibhai, M.F. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure, Mutagenesis, and Activity Studies Reveal that Patatin Is a
Lipid Acyl Hydrolase with a Ser-Asp Catalytic Dyad
Biochemistry, 42, 2003
|
|
3EYP
 
 | | Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Putative alpha-L-fucosidase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative alpha-L-fucosidase from Bacteroides thetaiotaomicron
To be Published
|
|
3F2B
 
 | | DNA Polymerase PolC from Geobacillus kaustophilus complex with DNA, dGTP, Mg and Zn | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*DAP*DTP*DAP*DAP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DTP*DCP*DAP*DCP*DTP*DG)-3', 5'-D(*DCP*DAP*DGP*DTP*DGP*DAP*DGP*DAP*DCP*DGP*DGP*DGP*DCP*DAP*DAP*DCP*DC)-3', ... | | Authors: | Davies, D.R, Evans, R.J, Bullard, J.M, Christensen, J, Green, L.S, Guiles, J.W, Ribble, W.K, Janjic, N, Jarvis, T.C. | | Deposit date: | 2008-10-29 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of PolC reveals unique DNA binding and fidelity determinants.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1PFP
 
 | | CATHELIN-LIKE MOTIF OF PROTEGRIN-3 | | Descriptor: | Protegrin 3 | | Authors: | Strub, M.-P, Hoh, F, Sanchez, J.-F, Strub, J.M, Bock, A, Aumelas, A, Dumas, C. | | Deposit date: | 2003-05-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing
Structure, 11, 2003
|
|
3F7P
 
 | |
1PIY
 
 | | RIBONUCLEOTIDE REDUCTASE R2 SOAKED WITH FERROUS ION AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PIM
 
 | | DITHIONITE REDUCED E. COLI RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT, D84E MUTANT | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
3EVW
 
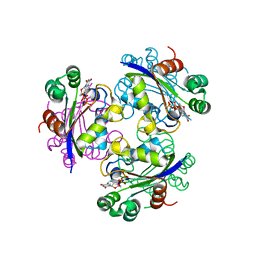 | | Crystal structure of the Mimivirus NDK R107G mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-10-13 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3F7Q
 
 | |
3FCW
 
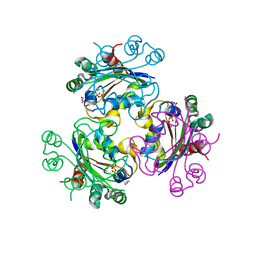 | | Crystal structure of the Mimivirus NDK N62L mutant complexed with UDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-23 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3EVY
 
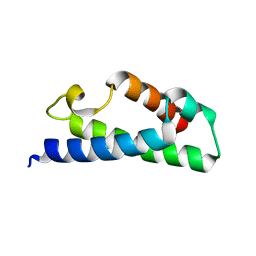 | | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis | | Descriptor: | Putative type I restriction enzyme R protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a fragment of a putative type I restriction enzyme R protein from Bacteroides fragilis
To be Published
|
|
3FMY
 
 | |
1PJ0
 
 | | RIBONUCLEOTIDE REDUCTASE R2-D84E/W48F MUTANT SOAKED WITH FERROUS IONS AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1PIZ
 
 | | RIBONUCLEOTIDE REDUCTASE R2 D84E MUTANT SOAKED WITH FERROUS IONS AT NEUTRAL PH | | Descriptor: | FE (III) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
3F13
 
 | | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum | | Descriptor: | putative nudix hydrolase family member | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum
To be Published
|
|
1PIU
 
 | | OXIDIZED RIBONUCLEOTIDE REDUCTASE R2-D84E MUTANT CONTAINING OXO-BRIDGED DIFERRIC CLUSTER | | Descriptor: | FE (III) ION, MERCURY (II) ION, OXYGEN ATOM, ... | | Authors: | Voegtli, W.C, Khidekel, N, Baldwin, J, Ley, B.A, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-05-30 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Ribonucleotide Reductase R2 Mutant that Accumulates a u-1,2-Peroxodiiron(III)
Intermediate during Oxygen Activation
J.Am.Chem.Soc., 122, 2000
|
|
3F1C
 
 | | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes | | Descriptor: | Putative 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase 2 | | Authors: | Patskovsky, Y, Ho, J, Toro, R, Gilmore, M, Miller, S, Groshong, C, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Listeria monocytogenes
To be Published
|
|
3F5S
 
 | | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
1PJV
 
 | | Cobatoxin 1 from Centruroides noxius Scorpion venom: Chemical Synthesis, 3-D Structure in Solution, Pharmacology and Docking on K+ channels | | Descriptor: | Cobatoxin 1 | | Authors: | Mosbah, A, Jouirou, B, Visan, V, Grissmer, S, El Ayeb, M, Rochat, H, De Waard, M, Mabrouk, K, Sabatier, J.M. | | Deposit date: | 2003-06-03 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Cobatoxin 1 from Centruroides noxius scorpion venom: chemical synthesis, three-dimensional structure in solution, pharmacology and docking on K+ channels.
Biochem.J., 377, 2004
|
|
