7B13
 
 | |
7AOF
 
 | | Atomic structure of the poxvirus transcription late pre-initiation complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6UYM
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v6 redesigned core from genotype 1a bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Zhu, J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
1GHK
 
 | | SOLUTION STRUCTURE OF THE LIPOYL DOMAIN OF THE 2-OXOGLUTARATE DEHYDROGENASE COMPLEX FROM AZOTOBACTER VINELAND II, NMR, 25 STRUCTURES | | Descriptor: | E2, THE DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE COMPONENT OF 2-OXOGLUTARATE DEHYDROGENASE COMPLEX | | Authors: | Berg, A, Vervoort, J, De Kok, A. | | Deposit date: | 1996-01-16 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lipoyl domain of the 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii.
J.Mol.Biol., 261, 1996
|
|
7SVU
 
 | | TnsBctd-TnsC-TniQ complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (28-MER), DNA (29-MER), ... | | Authors: | Park, J, Tsai, A.W.T, Kellogg, E.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the holo CRISPR RNA-guided transposon integration complex
Nature, 613, 2023
|
|
8RRR
 
 | | Alpha-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Saibil, H.R, Monistrol, J. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise recruitment of Hsc70 by DNAJB1 produces ordered arrays primed for bursts of amyloid fibre disassembly
Biorxiv, 2024
|
|
8RQM
 
 | | Alpha-synuclein amyloid fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Saibil, H.R, Monistrol, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise recruitment of Hsc70 by DNAJB1 produces ordered arrays primed for bursts of amyloid fibre disassembly
Biorxiv, 2024
|
|
7AFX
 
 | |
7AIE
 
 | |
8QDO
 
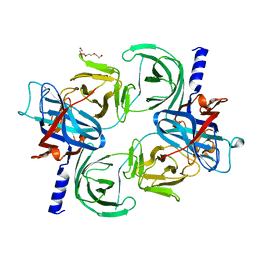 | | Crystal structure of the tegument protein UL82 (pp71) from Human Cytomegalovirus | | Descriptor: | Protein pp71, TETRAETHYLENE GLYCOL | | Authors: | Bresch, I.P, Eberhage, J, Reubold, T.F, Eschenburg, S. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tegument protein UL82 (pp71) from human cytomegalovirus.
Protein Sci., 33, 2024
|
|
6V15
 
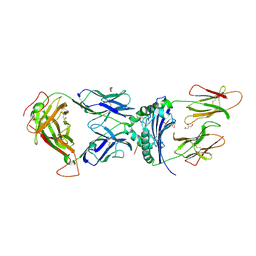 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, G08 TCR alpha chain, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
7DVM
 
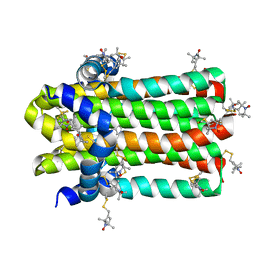 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
7AEZ
 
 | |
6UYF
 
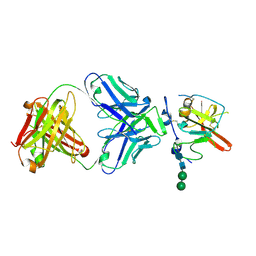 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v1 redesigned core from genotype 6a bound to broadly neutralizing antibody AR3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Zhu, J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
3JZH
 
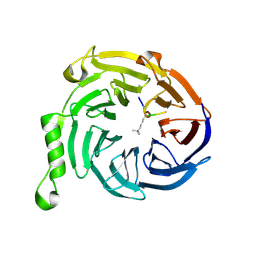 | | EED-H3K79me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7AOZ
 
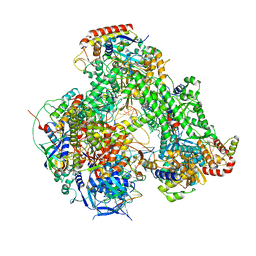 | | Atomic structure of the poxvirus transcription initiation complex in conformation 1 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP8
 
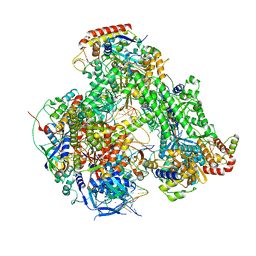 | | Atomic structure of the poxvirus initially transcribing complex in conformation 2 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6VFP
 
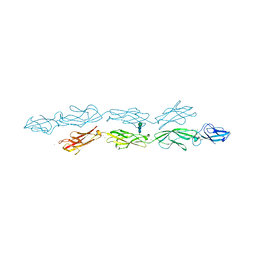 | | Crystal structure of human protocadherin 1 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin-1, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
6VFU
 
 | | Crystal structure of human protocadherin 19 EC1-EC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
7AFY
 
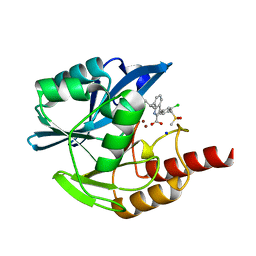 | | Crystal structure of the metallo-beta-lactamase VIM1 with 1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, SODIUM ION, ... | | Authors: | Brem, J, Schofield, C.J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.105 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM2 with 139
To Be Published
|
|
7AOH
 
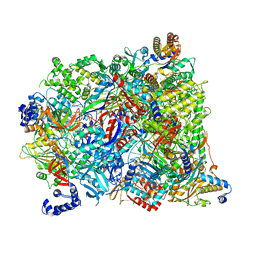 | | Atomic structure of the poxvirus late initially transcribing complex | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AP9
 
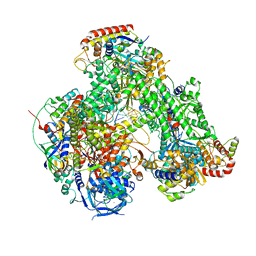 | | Atomic structure of the poxvirus initially transcribing complex in conformation 3 | | Descriptor: | DNA-directed RNA polymerase, DNA-directed RNA polymerase 147 kDa polypeptide, DNA-directed RNA polymerase 18 kDa subunit, ... | | Authors: | Grimm, C, Bartuli, J, Fischer, U. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of the complete poxvirus transcription initiation process.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8RFE
 
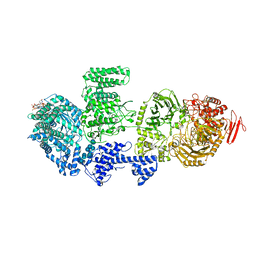 | | CgsiGP2 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, URIDINE-5'-DIPHOSPHATE-GLUCOSE, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
7AKQ
 
 | |
6V8Q
 
 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
