8QI9
 
 | | CrPhotLOV1 dark state structure determined by serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
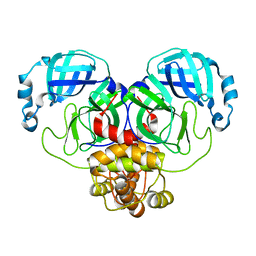
1A06
 
 | | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE | | Authors: | Kuriyan, J, Goldberg, J. | | Deposit date: | 1997-12-09 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I.
Cell(Cambridge,Mass.), 84, 1996
|
|
8QIN
 
 | | CrPhotLOV1 light state structure 47.5 ms (45-50 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
6Q7A
 
 | |
8QIP
 
 | | CrPhotLOV1 light state structure 57.5 ms (55-60 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
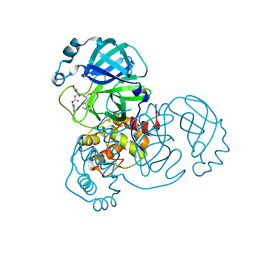
8D4L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
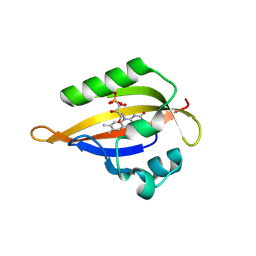
8D4M
 
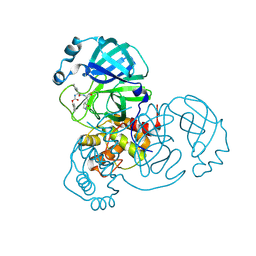 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8QI8
 
 | | Cryogenic temperature dark state structure of CrPhotLOV1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIA
 
 | | CrPhotLOV1 light state structure 2.5 ms (0-5 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIH
 
 | | CrPhotLOV1 light state structure 22.5 ms (20-25 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8DKY
 
 | |
8QIU
 
 | | CrPhotLOV1 light state structure 82.5 ms (80-85 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIL
 
 | | CrPhotLOV1 light state structure 37.5 ms (35-40 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIO
 
 | | CrPhotLOV1 light state structure 52.5 ms (50-55 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIR
 
 | | CrPhotLOV1 light state structure 67.5 ms (65-70 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8D4J
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8QIT
 
 | | CrPhotLOV1 light state structure 77.5 ms (75-80 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8D4N
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8QII
 
 | | CrPhotLOV1 light state structure 27.5 ms (25-30 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIQ
 
 | | CrPhotLOV1 light state structure 62.5 ms (60-65 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
8QIW
 
 | | CrPhotLOV1 light state structure 92.5 ms (90-95 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
6NKO
 
 | | Crystal structure of ForH | | Descriptor: | ForH | | Authors: | Zheng, J, Irani, S, Zhang, Y. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Identification of the Formycin A Biosynthetic Gene Cluster from Streptomyces kaniharaensis Illustrates the Interplay between Biological Pyrazolopyrimidine Formation and de Novo Purine Biosynthesis.
J. Am. Chem. Soc., 141, 2019
|
|
5KCU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
8QIB
 
 | | CrPhotLOV1 light state structure 7.5 ms (5-10 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-11 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography.
Iucrj, 2024
|
|
