1DRN
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GGAGA)R(UGAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*GP*AP*GP*A)-R(P*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-11-15 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
1DHH
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DUPLEX CONTAINING DNA/RNA HYBRID REGION, D(GG)R(AGAU)D(GAC)/D(GTCATCTCC) | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*CP*TP*CP*C)-3'), DNA/RNA (5'-D(*GP*G)-R(P*AP*GP*AP*U)-D(P*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohkubo, T, Kojima, C, Nakamura, H, Kyogoku, Y, Ohtsuka, E. | | Deposit date: | 1995-10-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Strucutres of DNA duplexes containing a DNA x RNA hybrid region, d(GG)r(AGAU)d(GAC) x d(GTCATCTCC) and d(GGAGA)r(UGAC) x d(GTCATCTCC).
Biochemistry, 35, 1996
|
|
1NAO
 
 | | SOLUTION STRUCTURE OF AN RNA 2'-O-METHYLATED RNA DUPLEX CONTAINING AN RNA/DNA HYBRID SEGMENT AT THE CENTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*OMGP*OMUP*OMC)-D(P*AP*TP*CP*T)-R(P*OMCP*OMC)-3'), RNA (5'-R(*GP*GP*AP*GP*AP*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohtsuka, E, Nakamura, H. | | Deposit date: | 1996-03-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA.2'-O-methylated RNA hybrid duplex containing an RNA.DNA hybrid segment at the center.
Biochemistry, 36, 1997
|
|
4YM5
 
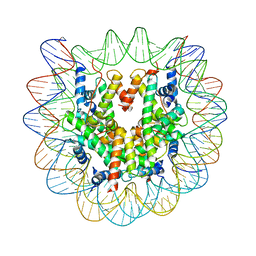 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4YM6
 
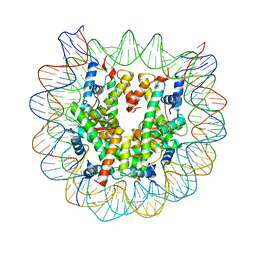 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
4GLE
 
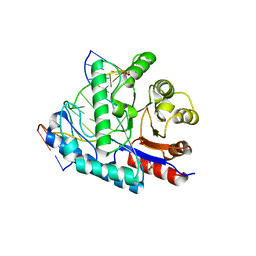 | | SacUVDE in complex with 6-4PP-containing DNA | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*CP*AP*AP*GP*GP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*CP*CP*(64T)P*(5PY)P*GP*AP*CP*GP*AP*CP*G)-3', SULFATE ION, ... | | Authors: | Meulenbroek, E.M, Peron Cane, C, Jala, I, Iwai, S, Moolenaar, G.F, Goosen, N, Pannu, N.S. | | Deposit date: | 2012-08-14 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions.
Nucleic Acids Res., 41, 2013
|
|
1VAS
 
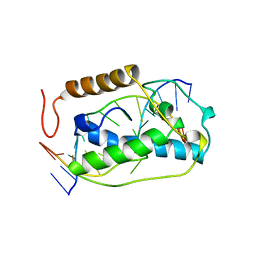 | | ATOMIC MODEL OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME COMPLEXED WITH A DNA SUBSTRATE: STRUCTURAL BASIS FOR DAMAGED DNA RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*CP*GP*TP*TP*GP*CP*GP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*GP*A)-3'), PROTEIN (T4 ENDONUCLEASE V (E.C.3.1.25.1)) | | Authors: | Vassylyev, D.G, Kashiwagi, T, Mikami, Y, Ariyoshi, M, Iwai, S, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1995-09-08 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition.
Cell(Cambridge,Mass.), 83, 1995
|
|
4A09
 
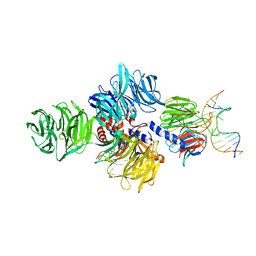 | | Structure of hsDDB1-drDDB2 bound to a 15 bp CPD-duplex (purine at D-1 position) at 3.1 A resolution (CPD 2) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*GP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0B
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A0A
 
 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.6 A resolution (CPD 3) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*GP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', CALCIUM ION, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
4A08
 
 | | Structure of hsDDB1-drDDB2 bound to a 13 bp CPD-duplex (purine at D-1 position) at 3.0 A resolution (CPD 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*CP*GP*CP*GP*AP*(TTD)P*GP*CP*GP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*CP*GP*CP*CP*CP*TP*CP*GP*CP*G)-3', ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
