5XCB
 
 | |
6IXZ
 
 | |
5YFK
 
 | |
6IXY
 
 | |
1IZE
 
 | | Crystal structure of Aspergillus oryzae Aspartic proteinase complexed with pepstatin | | 分子名称: | Pepstatin, alpha-D-mannopyranose, aspartic proteinase | | 著者 | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | 登録日 | 2002-10-02 | | 公開日 | 2003-03-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of Aspergillus oryzae aspartic proteinase and its complex with an inhibitor pepstatin at 1.9A resolution.
J.Mol.Biol., 326, 2003
|
|
1IZD
 
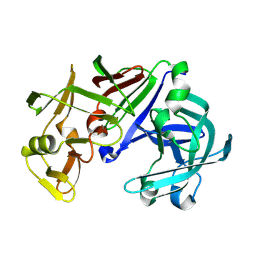 | | Crystal structure of Aspergillus oryzae Aspartic Proteinase | | 分子名称: | Aspartic proteinase, alpha-D-mannopyranose | | 著者 | Kamitori, S, Ohtaki, A, Ino, H, Takeuchi, M. | | 登録日 | 2002-10-02 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Aspergillus oryzae Aspartic Proteinase and its Complex with an Inhibitor Pepstatin at 1.9 A Resolution
J.Mol.Biol., 326, 2003
|
|
7WRR
 
 | |
7WRT
 
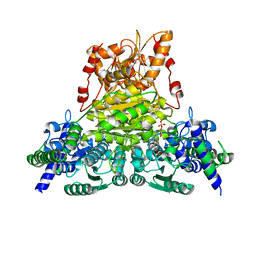 | |
2EIS
 
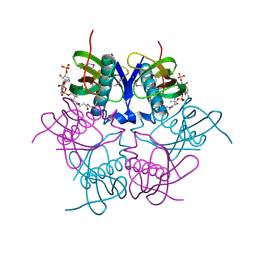 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | 分子名称: | COENZYME A, Hypothetical protein TTHB207 | | 著者 | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-13 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
6L68
 
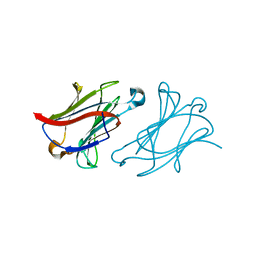 | | X-ray structure of human galectin-10 in complex with D-allose | | 分子名称: | Galectin-10, beta-D-allopyranose | | 著者 | Kamitori, S. | | 登録日 | 2019-10-28 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structures of human galectin-10/monosaccharide complexes demonstrate potential of monosaccharides as effectors in forming Charcot-Leyden crystals.
Biochem.Biophys.Res.Commun., 2020
|
|
6L6D
 
 | |
2D58
 
 | | Human microglia-specific protein Iba1 | | 分子名称: | Allograft inflammatory factor 1, NICKEL (II) ION | | 著者 | Kamitori, S. | | 登録日 | 2005-10-31 | | 公開日 | 2006-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-ray Structures of the Microglia/Macrophage-specific Protein Iba1 from Human and Mouse Demonstrate Novel Molecular Conformation Change Induced by Calcium binding
J.Mol.Biol., 364, 2006
|
|
3AZ1
 
 | | Crystal Structure Analysis of Vitamin D receptor | | 分子名称: | Vitamin D3 receptor, {4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetic acid | | 著者 | Itoh, S, Iijima, S. | | 登録日 | 2011-05-20 | | 公開日 | 2011-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ2
 
 | | Crystal Structure Analysis of Vitamin D receptor | | 分子名称: | 5-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}pentanoic acid, Vitamin D3 receptor | | 著者 | Itoh, S, Iijima, S. | | 登録日 | 2011-05-20 | | 公開日 | 2011-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AZ3
 
 | | Crystal Structure Analysis of Vitamin D receptor | | 分子名称: | (4S)-4-hydroxy-5-[4-(3-{4-[(3S)-3-hydroxy-4,4-dimethylpentyl]-3-methylphenyl}pentan-3-yl)-2-methylphenoxy]pentanoic acid, Vitamin D3 receptor | | 著者 | Itoh, S, Iijima, S. | | 登録日 | 2011-05-20 | | 公開日 | 2011-11-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
7D6P
 
 | |
7D6T
 
 | |
7CFL
 
 | |
7F5I
 
 | | X-ray structure of Clostridium perfringens-specific amidase endolysin | | 分子名称: | GLUTAMIC ACID, SODIUM ION, ZINC ION, ... | | 著者 | Kamitori, S, Tamai, E. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and biochemical characterization of the Clostridium perfringens-specific Zn 2+ -dependent amidase endolysin, Psa, catalytic domain.
Biochem.Biophys.Res.Commun., 576, 2021
|
|
3B4X
 
 | |
3VKN
 
 | |
3VKL
 
 | |
3VKO
 
 | | Galectin-8 N-terminal domain in complex with sialyllactosamine | | 分子名称: | CHLORIDE ION, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Kamitori, S, Yoshida, H. | | 登録日 | 2011-11-18 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3W0Y
 
 | | Crystal Structure Analysis of Vitamin D receptor | | 分子名称: | Vitamin D3 receptor, [3-fluoro-2'-methyl-4'-(3-{3-methyl-4-[(1E)-4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-en-1-yl]phenyl}pentan-3-yl)biphenyl-4-yl]acetic acid | | 著者 | Itoh, S, Iijima, S. | | 登録日 | 2012-11-05 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal Structure Analysis of Vitamin D receptor
TO BE PUBLISHED
|
|
3W0C
 
 | | Crystal Structure Analysis of Vitamin D receptor | | 分子名称: | (4S)-4-hydroxy-5-[2-methyl-4-(3-{3-methyl-4-[(1E)-4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-en-1-yl]phenyl}pentan-3-yl)phenoxy]pentanoic acid, Vitamin D3 receptor | | 著者 | Itoh, S, Iijima, S. | | 登録日 | 2012-10-29 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | structure analysis of vitamin D receptor
To be Published
|
|
