8AGM
 
 | | Limonene epoxide low pH soak of epoxide hydrolase from metagenomic source ch65 | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, CHLORIDE ION, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8AGP
 
 | | Halogenated product of limonene epoxide turnover by epoxide hydrolase from metagenomic source ch65 | | Descriptor: | (1~{S},2~{S},4~{R})-2-chloranyl-1-methyl-4-prop-1-en-2-yl-cyclohexan-1-ol, 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8AXG
 
 | | Crystal structure of Fusobacterium nucleatum fusolisin protease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fusolisin, ... | | Authors: | Isupov, M.N, Wiener, R, Rouvinski, A, Fahoum, J, Kumar, M, Read, R.J. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of Fusobacterium nucleatum fusolisin protease
To Be Published
|
|
7PNB
 
 | | Sulfolobus acidocaldarius 0406 filament. | | Descriptor: | 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sulfolobus acidocaldarius 0406 filament., beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Isupov, M.N, Gaines, M, Daum, B. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Electron cryo-microscopy reveals the structure of the archaeal thread filament.
Nat Commun, 13, 2022
|
|
7Q9X
 
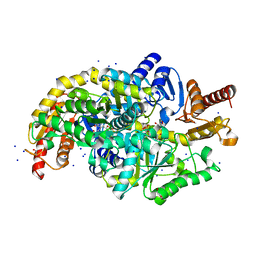 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
7Q9Z
 
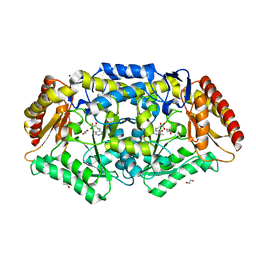 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
7QW3
 
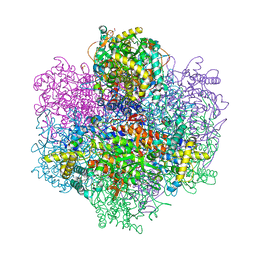 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with Br ion. | | Descriptor: | BROMIDE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QVW
 
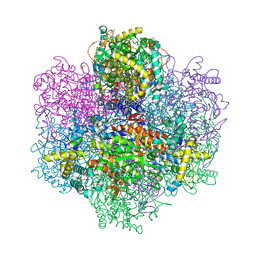 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QWI
 
 | | Vanadate complex of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QYY
 
 | | Vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with chloride | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
4UT4
 
 | | Burkholderia pseudomallei heptokinase WcbL, D-mannose complex. | | Descriptor: | CHLORIDE ION, PUTATIVE SUGAR KINASE, alpha-D-mannopyranose | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
7ZCX
 
 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-03-29 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
5AIF
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample-Native | | Descriptor: | IMIDAZOLE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
5I6J
 
 | |
5I7D
 
 | |
1W5T
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADPNP-ADP complexes) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ORC2, ... | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
1W5S
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADP form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ORIGIN RECOGNITION COMPLEX SUBUNIT 2 ORC2, SULFATE ION | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
5I6R
 
 | | Crystal Structure of srGAP2 F-BARx WT Form-1 | | Descriptor: | ACETATE ION, D-MALATE, SLIT-ROBO Rho GTPase-activating protein 2, ... | | Authors: | Sporny, M, Guez-Haddad, J, Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2016-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for srGAP2 Membrane Interactions, and Antagonism by the Human Specific Paralog srGAP2C
To Be Published
|
|
4USK
 
 | | Unravelling the B. pseudomallei heptokinase WcbL: from Structure to Drug Discovery. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, PUTATIVE SUGAR KINASE, ... | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4UTG
 
 | | Burkholderia pseudomallei heptokinase WcbL,AMPPNP (ATP analogue) complex. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4USM
 
 | | WcbL complex with glycerol bound to sugar site | | Descriptor: | CHLORIDE ION, GLYCEROL, PUTATIVE SUGAR KINASE | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
1H2B
 
 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2002-08-02 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
5OSI
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL harpin loop (163-176) | | Descriptor: | 1,2-ETHANEDIOL, Interaptin, SODIUM ION, ... | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OSH
 
 | | Structure of retromer VPS29-VPS35C subunits complexed with RidL N-terminal domain (1-236) | | Descriptor: | Interaptin, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OT4
 
 | | Structure of the Legionella pneumophila effector RidL (1-866) | | Descriptor: | GLYCEROL, Interaptin | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
