3VSD
 
 | | Crystal Structure of the K127A Mutant of O-Phosphoserine Sulfhydrylase Complexed with External Schiff Base of Pyridoxal 5'-Phosphate with O-Acetyl-L-Serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, O-ACETYLSERINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
3VSC
 
 | | Crystal Structure of the K127A Mutant of O-Phosphoserine Sulfhydrylase Complexed with External Schiff Base of Pyridoxal 5'-Phosphate with O-Phospho-L-Serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOSERINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
3AI7
 
 | | Crystal Structure of Bifidobacterium Longum Phosphoketolase | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, Xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | | Authors: | Takahashi, K, Tagami, U, Shimba, N, Kashiwagi, T, Ishikawa, K, Suzuki, E. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Bifidobacterium Longum phosphoketolase; key enzyme for glucose metabolism in Bifidobacterium
Febs Lett., 584, 2010
|
|
3A4W
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, MAGNESIUM ION, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
2ZUN
 
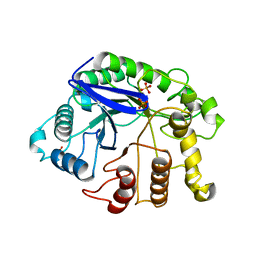 | |
3A2W
 
 | | Peroxiredoxin (C50S) from Aeropytum pernix K1 (peroxide-bound form) | | Descriptor: | GLYCEROL, PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A2V
 
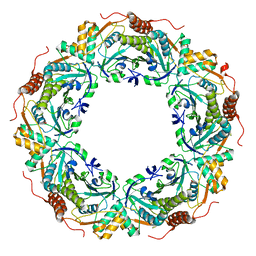 | | Peroxiredoxin (C207S) from Aeropyrum pernix K1 complexed with hydrogen peroxide | | Descriptor: | PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Ishikawa, K, Matsumura, H, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3A5W
 
 | | Peroxiredoxin (wild type) from Aeropyrum pernix K1 (reduced form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, T, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
2ZUM
 
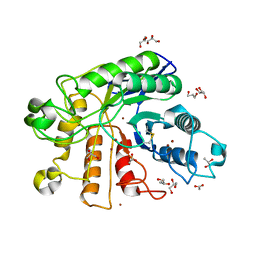 | |
3A2X
 
 | | Peroxiredoxin (C50S) from Aeropyrum pernix K1 (acetate-bound form) | | Descriptor: | ACETATE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3APG
 
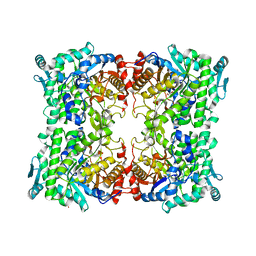 | |
2Z6N
 
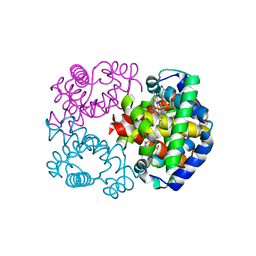 | | Crystal Structure of Carbonmonoxy Hemoglobin D from the Aldabra Giant Tortoise, Geochelone gigantea | | Descriptor: | CARBON MONOXIDE, Hemoglobin A/D subunit beta, Hemoglobin D subunit alpha, ... | | Authors: | Kuwada, T, Hasegawa, T, Sato, I, Ishikawa, K, Shishikura, F. | | Deposit date: | 2007-08-04 | | Release date: | 2007-08-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Ligated Hemoglobin D from the Aldabra Giant Tortoise, Geochelone gigantea
To be Published
|
|
3VGI
 
 | | The crystal structure of hyperthermophilic family 12 endocellulase from Pyrococcus furiosus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Endoglucanase A, ... | | Authors: | Kataoka, M, Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-08-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic resolution of the crystal structure of the hyperthermophilic family 12 endocellulase and stabilizing role of the DxDxDG calcium-binding motif in Pyrococcus furiosus.
Febs Lett., 586, 2012
|
|
3WDP
 
 | | Structural analysis of a beta-glucosidase mutant derived from a hyperthermophilic tetrameric structure | | Descriptor: | Beta-glucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Nakabayashi, M, Kataoka, M, Ishikawa, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of beta-glucosidase mutants derived from a hyperthermophilic tetrameric structure.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WQ1
 
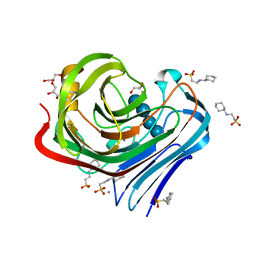 | |
3WQ7
 
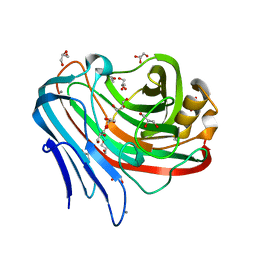 | |
3WQ8
 
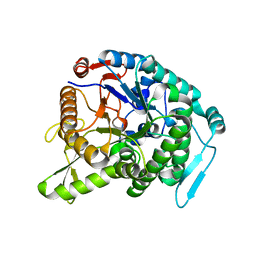 | | Monomer structure of hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form | | Descriptor: | Beta-glucosidase | | Authors: | Nakabayashi, M, Kataoka, M, Watanabe, M, Ishikawa, K. | | Deposit date: | 2014-01-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3W6M
 
 | |
3W6L
 
 | |
3WT3
 
 | |
3WY6
 
 | |
3WQ0
 
 | |
3WR0
 
 | |
3WXP
 
 | |
3WO8
 
 | | Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | Beta-N-acetylglucosaminidase | | Authors: | Mine, S, Kado, Y, Watanabe, M, Inoue, T, Ishikawa, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The structure of hyperthermophilic beta-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site.
Febs J., 281, 2014
|
|
