5X5I
 
 | | The X-ray crystal structure of a TetR family transcription regulator RcdA involved in the regulation of biofilm formation in Escherichia coli | | Descriptor: | HTH-type transcriptional regulator RcdA | | Authors: | Sugino, H, Usui, M, Shimada, T, Nakano, M, Ogasawara, H, Ishihama, A, Hirata, A. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A structural sketch of RcdA, a transcription factor controlling the master regulator of biofilm formation.
FEBS Lett., 591, 2017
|
|
7VVI
 
 | | OXA-58 crystal structure of acylated meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, SULFATE ION | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex
to be published
|
|
7VX3
 
 | | OXA-58 crystal structure of acylated meropenem complex 2 | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, ... | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex 2
To be published
|
|
7VX6
 
 | | OXA-58 crystal structure of acylated meropenem complex 2 | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, ... | | Authors: | Saino, H, Sugiyabu, T, Miyano, M. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | OXA-58 crystal structure of acylated meropenem complex 2
To be published
|
|
2Z07
 
 | |
5BOH
 
 | |
2Z08
 
 | |
2Z09
 
 | |
2Z3V
 
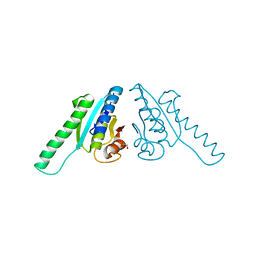 | |
3B29
 
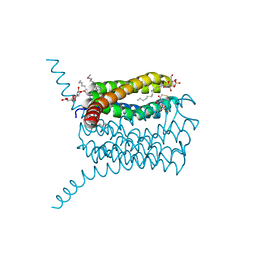 | | Human leukotriene C4 synthase in complex with dodecyl-beta-D-selenomaltoside | | Descriptor: | GLUTATHIONE, Leukotriene C4 synthase, SULFATE ION, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2011-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Seleno-detergent MAD phasing of leukotriene C4 synthase in complex with dodecyl-beta-D-selenomaltoside
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1WPK
 
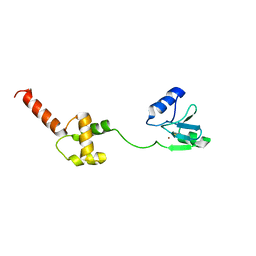 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | Descriptor: | ADA regulatory protein, ZINC ION | | Authors: | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2004-09-07 | | Release date: | 2005-09-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
1WJG
 
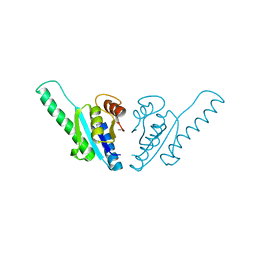 | |
1GEA
 
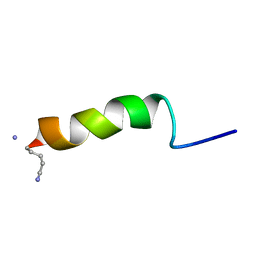 | | RECEPTOR-BOUND CONFORMATION OF PACAP21 | | Descriptor: | PITUITARY ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE | | Authors: | Inooka, H, Ohtaki, T, Kitahara, O, Ikegami, T, Endo, S, Kitada, C, Ogi, K, Onda, H, Fujino, M, Shirakawa, M. | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Conformation of a peptide ligand bound to its G-protein coupled receptor.
Nat.Struct.Biol., 8, 2001
|
|
3PCV
 
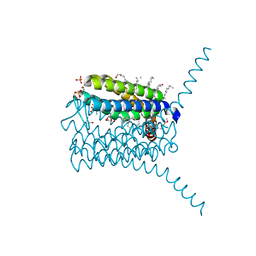 | | Crystal structure analysis of human leukotriene C4 synthase at 1.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The catalytic architecture of leukotriene C4 synthase with two arginine residues
J.Biol.Chem., 286, 2011
|
|
3WQ4
 
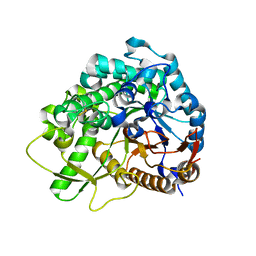 | | Crystal structure of beta-primeverosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-primeverosidase | | Authors: | Saino, H. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of beta-primeverosidase in complex with disaccharide amidine inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WQ5
 
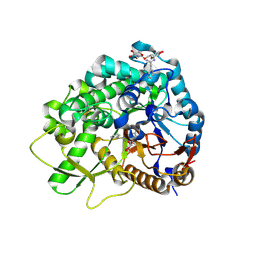 | |
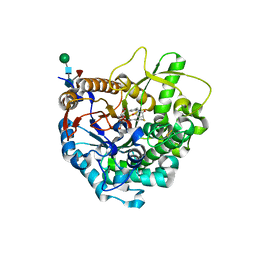
3WQ6
 
 | |
2CVE
 
 | |
4V0I
 
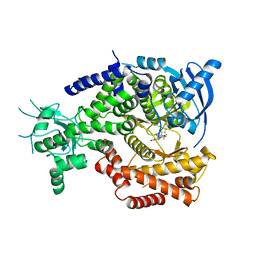 | | Water Network Determines Selectivity for a Series of Pyrimidone Indoline Amide PI3KBeta Inhibitors over PI3K-Delta | | Descriptor: | 2-[2-(2-METHYL-2,3-DIHYDRO-INDOL-1-YL)-2-OXO-ETHYL]-6-MORPHOLIN-4-YL-3H-PYRIMIDIN-4-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Robinson, D, Bertrand, T, Carry, J.C, Halley, F, Karlsson, A, Mathieu, M, Minoux, H, Perrin, M.A, Robert, B, Schio, L, Sherman, W. | | Deposit date: | 2014-09-16 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Differential Water Thermodynamics Determine Pi3K-Beta/Delta Selectivity for Solvent-Exposed Ligand Modifications.
J.Chem.Inf.Model., 56, 2016
|
|
1A0F
 
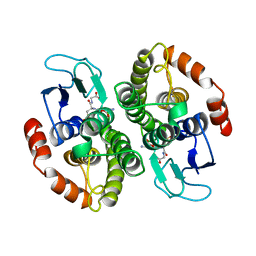 | | CRYSTAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE FROM ESCHERICHIA COLI COMPLEXED WITH GLUTATHIONESULFONIC ACID | | Descriptor: | GLUTATHIONE S-TRANSFERASE, GLUTATHIONE SULFONIC ACID | | Authors: | Nishida, M, Harada, S, Noguchi, S, Inoue, H, Takahashi, K, Satow, Y. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of Escherichia coli glutathione S-transferase complexed with glutathione sulfonate: catalytic roles of Cys10 and His106.
J.Mol.Biol., 281, 1998
|
|
4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
3X05
 
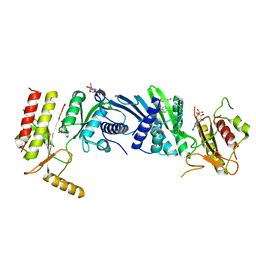 | | Crystal structure of PIP4KIIBETA T201M complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X06
 
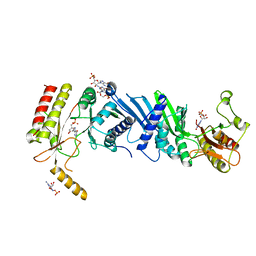 | | Crystal structure of PIP4KIIBETA T201M complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X01
 
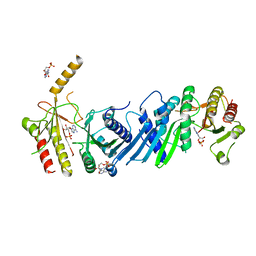 | | Crystal structure of PIP4KIIBETA complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X09
 
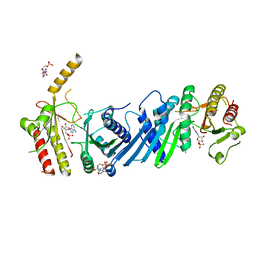 | | Crystal structure of PIP4KIIBETA F205L complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
