2B7N
 
 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori | | Descriptor: | Probable nicotinate-nucleotide pyrophosphorylase, QUINOLINIC ACID, SULFATE ION | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-04 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
2B8I
 
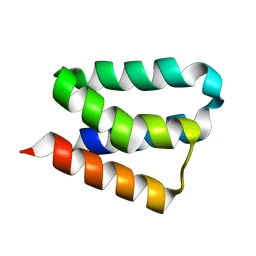 | | Crystal Structure and Functional Studies Reveal that PAS Factor from Vibrio vulnificus is a Novel Member of the Saposin-Fold Family | | Descriptor: | PAS factor | | Authors: | Lee, J.H, Yang, S.T, Rho, S.H, Im, Y.J, Kim, S.Y, Kim, Y.R, Kim, M.K, Kang, G.B, Kim, J.I, Rhee, J.H, Eom, S.H. | | Deposit date: | 2005-10-07 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and functional studies reveal that PAS factor from Vibrio vulnificus is a novel member of the saposin-fold family
J.Mol.Biol., 355, 2006
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
2B7Q
 
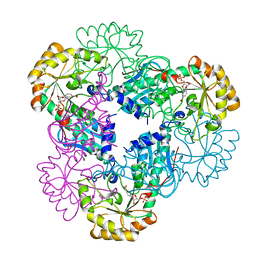 | | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori with nicotinate mononucleotide | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Probable nicotinate-nucleotide pyrophosphorylase | | Authors: | Kim, M.K, Im, Y.J, Lee, J.H, Eom, S.H. | | Deposit date: | 2005-10-05 | | Release date: | 2006-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori
Proteins, 63, 2006
|
|
1XNG
 
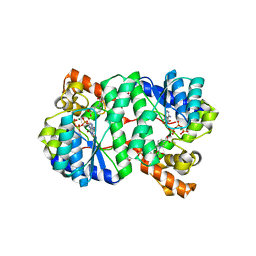 | | Crystal Structure of NH3-dependent NAD+ synthetase from Helicobacter pylori | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, ... | | Authors: | Kang, G.B, Kim, Y.S, Im, Y.J, Rho, S.H, Lee, J.H, Eom, S.H. | | Deposit date: | 2004-10-05 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Helicobacter pylori
Proteins, 58, 2005
|
|
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease from Vibrio vunificus
To Be Published
|
|
8YJA
 
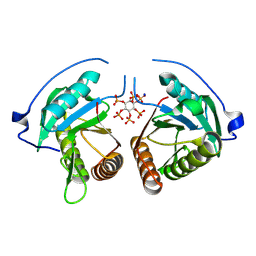 | | Structure of Vibrio vulnificus MARTX cysteine protease domain lacking beta-flap | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MARTX cysteine protease domain, SODIUM ION | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease from Vibrio vunificus
To Be Published
|
|
4J7Q
 
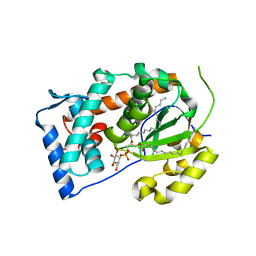 | | Crystal structure of Saccharomyces cerevisiae Sfh3 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
2IE8
 
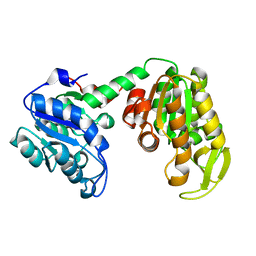 | |
4J7P
 
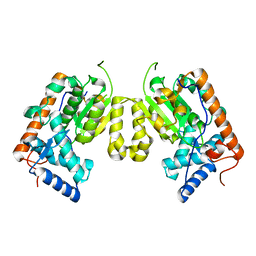 | | Crystal structure of Saccharomyces cerevisiae Sfh3 | | Descriptor: | Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
7VPR
 
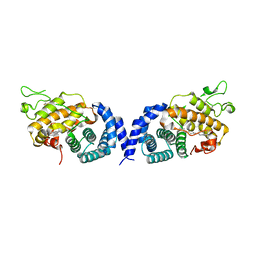 | |
7VPU
 
 | |
7VPS
 
 | |
7VPT
 
 | |
6L1D
 
 | |
6L1M
 
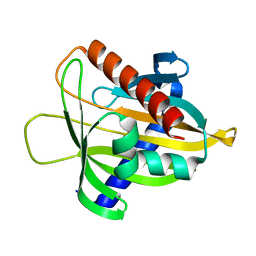 | |
5YQQ
 
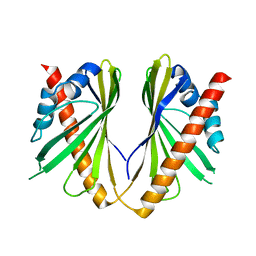 | |
7DEI
 
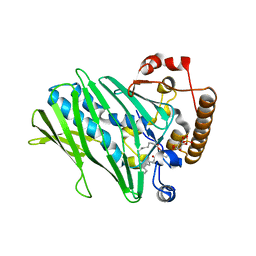 | | Structure of human ORP3 ORD domain in complex with PI(4)P | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Oxysterol-binding protein-related protein 3 | | Authors: | Tong, J, Tan, L, Im, Y.J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human ORP3 ORD reveals conservation of a key function and ligand specificity in OSBP-related proteins.
Plos One, 16, 2021
|
|
7DEJ
 
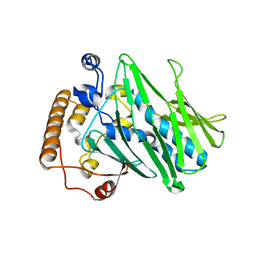 | | Structure of human ORP3 ORD in apo-form | | Descriptor: | Oxysterol-binding protein-related protein 3 | | Authors: | Tong, J, Tan, L, Im, Y.J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human ORP3 ORD reveals conservation of a key function and ligand specificity in OSBP-related proteins.
Plos One, 16, 2021
|
|
7F6J
 
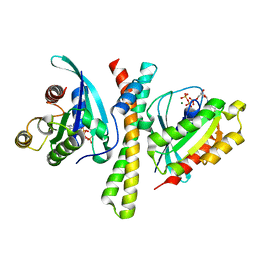 | | Crystal structure of the PDZD8 coiled-coil domain - Rab7 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PDZ domain-containing protein 8, ... | | Authors: | Khan, H, Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2021-06-25 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of human PDZD8-Rab7 interaction for the ER-late endosome tethering.
Sci Rep, 11, 2021
|
|
5YQI
 
 | | Crystal structure of the first StARkin domain of Lam2 | | Descriptor: | Membrane-anchored lipid-binding protein YSP2 | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YQP
 
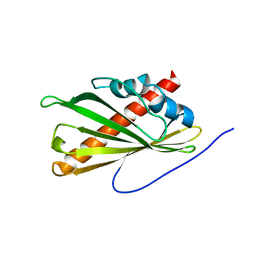 | | Crystal structure of the second StARkin domain of Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Tong, J, Manik, K.M, IM, Y.J. | | Deposit date: | 2017-11-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YS0
 
 | |
5YQR
 
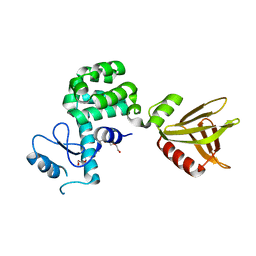 | | Crystal structure of the PH-like domain of Lam6 | | Descriptor: | Endolysin/Membrane-anchored lipid-binding protein LAM6 fusion protein, NONAETHYLENE GLYCOL | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2017-11-07 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7XB5
 
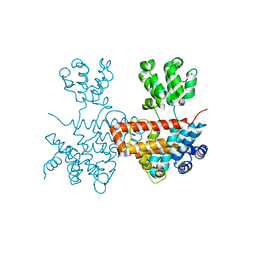 | |
