1WXI
 
 | | E.coli NAD Synthetase, AMP.PP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXH
 
 | | E.coli NAD Synthetase, NAD | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXE
 
 | | E.coli NAD Synthetase, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXF
 
 | | E.coli NAD Synthetase | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1Z1W
 
 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1N5W
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Oxidized form | | Descriptor: | CU(I)-S-MO(VI)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-07 | | Release date: | 2002-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N63
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Carbon monoxide reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N61
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Dithionite reduced state | | Descriptor: | CU(I)-S-MO(IV)(=O)OH CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N62
 
 | | Crystal Structure of the Mo,Cu-CO Dehydrogenase (CODH), n-butylisocyanide-bound state | | Descriptor: | CU(I)-S-MO(IV)(=O)O-NBIC CLUSTER, Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1N60
 
 | | Crystal Structure of the Cu,Mo-CO Dehydrogenase (CODH); Cyanide-inactivated Form | | Descriptor: | Carbon monoxide dehydrogenase large chain, Carbon monoxide dehydrogenase medium chain, Carbon monoxide dehydrogenase small chain, ... | | Authors: | Dobbek, H, Gremer, L, Kiefersauer, R, Huber, R, Meyer, O. | | Deposit date: | 2002-11-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Z5H
 
 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
1STC
 
 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1DMG
 
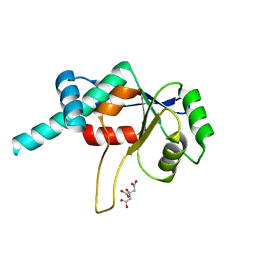 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L4 | | Descriptor: | CITRIC ACID, RIBOSOMAL PROTEIN L4 | | Authors: | Worbs, M, Huber, R, Wahl, M.C. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon.
EMBO J., 19, 2000
|
|
2NSM
 
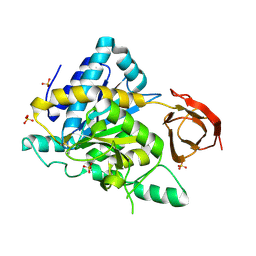 | | Crystal structure of the human carboxypeptidase N (Kininase I) catalytic domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase N catalytic chain, SULFATE ION | | Authors: | Keil, C, Maskos, K, Than, M, Hoopes, J.T, Huber, R, Tan, F, Deddish, P.A, Erdoes, E.G, Skidgel, R.A, Bode, W. | | Deposit date: | 2006-11-05 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain
J.Mol.Biol., 366, 2007
|
|
1ELQ
 
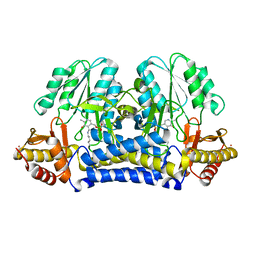 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ECX
 
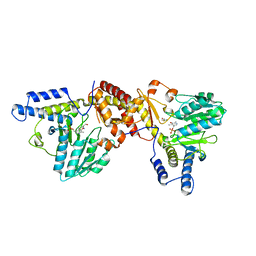 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kaiser, J.T, Clausen, T.C, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1EG5
 
 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1ELU
 
 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1JAE
 
 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Strobl, S, Maskos, K, Betz, M, Wiegand, G, Huber, R, Gomis-Rueth, F.X, Frank, G, Glockshuber, R. | | Deposit date: | 1997-09-30 | | Release date: | 1998-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yellow meal worm alpha-amylase at 1.64 A resolution.
J.Mol.Biol., 278, 1998
|
|
1PIF
 
 | | PIG ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Machius, M, Vertesy, L, Huber, R, Wiegand, G. | | Deposit date: | 1996-06-15 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carbohydrate and protein-based inhibitors of porcine pancreatic alpha-amylase: structure analysis and comparison of their binding characteristics.
J.Mol.Biol., 260, 1996
|
|
1PIG
 
 | | PIG PANCREATIC ALPHA-AMYLASE COMPLEXED WITH THE OLIGOSACCHARIDE V-1532 | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, ... | | Authors: | Machius, M, Vertesy, L, Huber, R, Wiegand, G. | | Deposit date: | 1996-06-15 | | Release date: | 1996-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbohydrate and protein-based inhibitors of porcine pancreatic alpha-amylase: structure analysis and comparison of their binding characteristics.
J.Mol.Biol., 260, 1996
|
|
1JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | BETA-MERCAPTOETHANOL, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-22 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1FS8
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES-SULFATE COMPLEX | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FIU
 
 | | TETRAMERIC RESTRICTION ENDONUCLEASE NGOMIV IN COMPLEX WITH CLEAVED DNA | | Descriptor: | ACETIC ACID, DNA (5'-D(*TP*GP*CP*G)-3'), DNA (5'-D(P*CP*CP*GP*GP*CP*GP*C)-3'), ... | | Authors: | Deibert, M, Grazulis, S, Sasnauskas, G, Siksnys, V, Huber, R. | | Deposit date: | 2000-08-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA.
Nat.Struct.Biol., 7, 2000
|
|
1FS7
 
 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
