8XL0
 
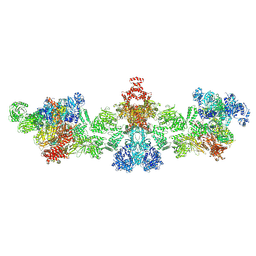 | | Citrate-induced filament of human acetyl-coenzyme A carboxylase 1 (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XKZ
 
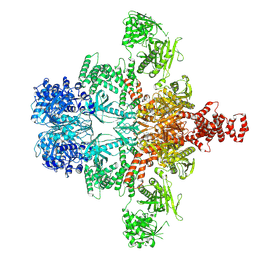 | | Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8TZS
 
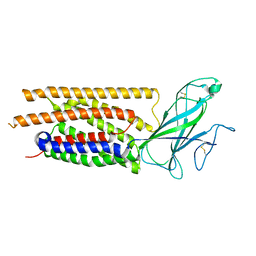 | | Structure of human WLS | | Descriptor: | Protein wntless homolog | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZP
 
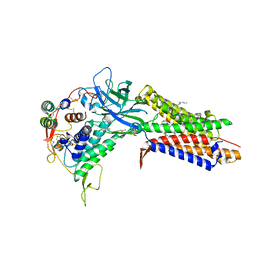 | | Structure of human Wnt7a bound to WLS and RECK | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PALMITOLEIC ACID, Protein Wnt-7a, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZR
 
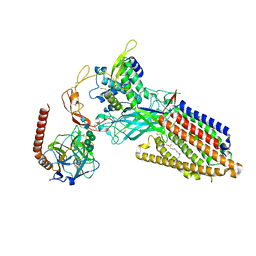 | | Structure of human Wnt3a bound to WLS and CALR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Calreticulin, PALMITOLEIC ACID, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
8TZO
 
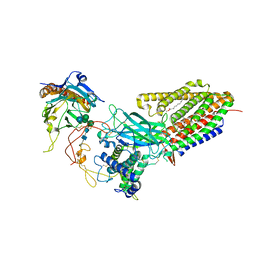 | | Structure of human Wnt7a bound to WLS and CALR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, Calreticulin, ... | | Authors: | Qi, X, Hu, Q, Li, X. | | Deposit date: | 2023-08-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Wnt biogenesis, secretion, and Wnt7-specific signaling.
Cell, 186, 2023
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
6CZ5
 
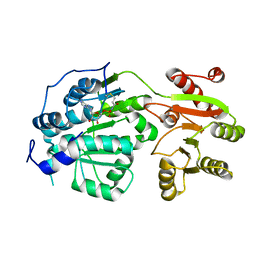 | | Crystal structure of small molecule AMP-acrylamide covalently bound to DDX3 S228C | | Descriptor: | 5'-O-[(R)-hydroxy(propanoylamino)phosphoryl]adenosine, ATP-dependent RNA helicase DDX3X | | Authors: | Barkovich, K.J, Moore, M.K, Hu, Q, Shokat, K.M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemical genetic inhibition of DEAD-box proteins using covalent complementarity.
Nucleic Acids Res., 46, 2018
|
|
8HF3
 
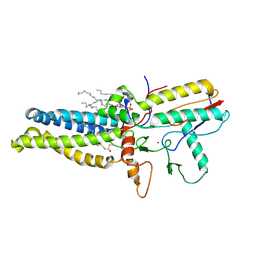 | | Cryo-EM structure of human ZDHHC9/GCP16 complex | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Golgin subfamily A member 7, PALMITIC ACID, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Liu, S, Yang, A. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HFC
 
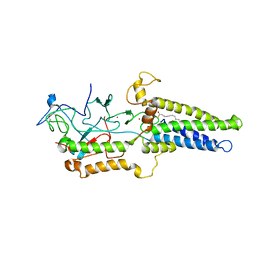 | | Cryo-EM structure of yeast Erf2/Erf4 complex | | Descriptor: | PALMITIC ACID, Palmitoyltransferase ERF2, Ras modification protein ERF4, ... | | Authors: | Wu, J, Hu, Q, Zhang, Y, Yang, A, Liu, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7XP9
 
 | | Phytophthora infesfans RxLR effector AVRvnt1 | | Descriptor: | RxLR effector protein Avr-vnt11 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Chloroplast Protein GLYK Hijacked by Phytophthora Infestans Effector AVRvnt1 in Cytoplasm to Activate NLR
To Be Published
|
|
5UMR
 
 | | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1 | | Descriptor: | FACT complex subunit SSRP1 | | Authors: | Su, D, Hu, Q, Thompson, J.R, Heroux, A, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SSRP1
To Be Published
|
|
5UMS
 
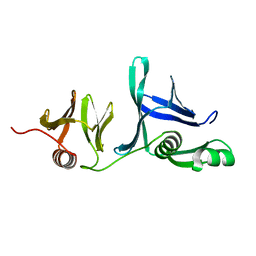 | |
5UMT
 
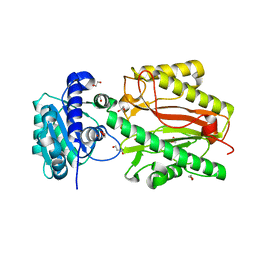 | | Crystal structure of N-terminal domain of human FACT complex subunit SPT16 | | Descriptor: | 1,2-ETHANEDIOL, FACT complex subunit SPT16, GLYCEROL | | Authors: | Su, D, Hu, Q, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2017-01-29 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Crystal structure of N-terminal domain of human FACT complex subunit SPT16
To Be Published
|
|
7C96
 
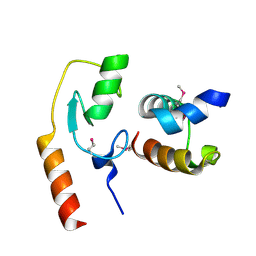 | | Avr1d:GmPUB13 U-box | | Descriptor: | RING-type E3 ubiquitin transferase, RxLR effector protein Avh6 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
7E40
 
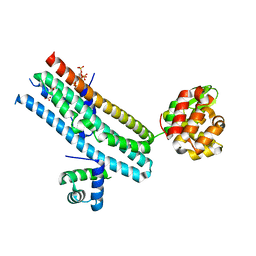 | | Mechanism of Phosphate Sensing and Signaling Revealed by Rice SPX1-PHR2 Complex Structure | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 1,Endolysin | | Authors: | Zhou, J, Hu, Q, Yao, D, Xing, W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure.
Nat Commun, 12, 2021
|
|
5H9T
 
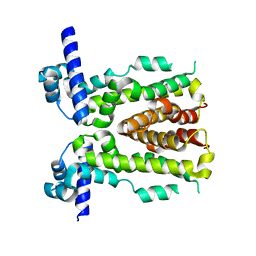 | | Crystal structure of native NalD at resolution of 2.9, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa.
Mol. Microbiol., 100, 2016
|
|
5DAJ
 
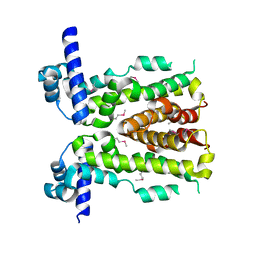 | | Crystal structure of NalD, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa
Mol.Microbiol., 100, 2016
|
|
2OMJ
 
 | | solution structure of LARG PDZ domain | | Descriptor: | Rho guanine nucleotide exchange factor 12 | | Authors: | Liu, J, Huang, H, Hu, Q. | | Deposit date: | 2007-01-22 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | solution structure and dynamics of the LARG PDZ domain
To be Published
|
|
