1UI6
 
 | |
1UI5
 
 | |
1ET8
 
 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASN MUTANT FROM ALCALIGENES FAECALIS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
3OIT
 
 | | Crystal structure of curcuminoid synthase CUS from Oryza sativa | | Descriptor: | Os07g0271500 protein | | Authors: | Miyazono, K, Um, J, Imai, F.L, Katsuyama, Y, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of curcuminoid synthase CUS from Oryza sativa
Proteins, 79, 2011
|
|
3OV3
 
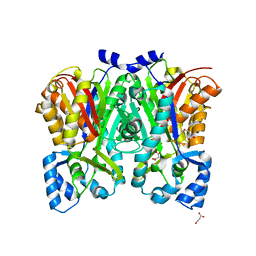 | | G211F mutant of curcumin synthase 1 from Curcuma longa | | Descriptor: | Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
1ET7
 
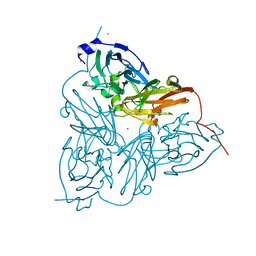 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASP MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | CADMIUM ION, COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
1ET5
 
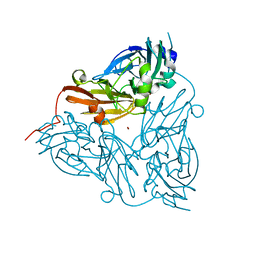 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE ASP98ASN MUTANT FROM ALCALIGENES FAECALIS S-6 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | Authors: | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | Deposit date: | 2000-04-12 | | Release date: | 2000-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
3OV2
 
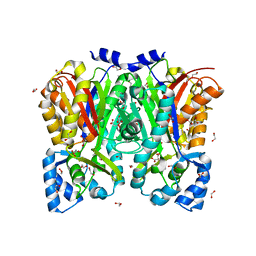 | | Curcumin synthase 1 from Curcuma longa | | Descriptor: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
3VS8
 
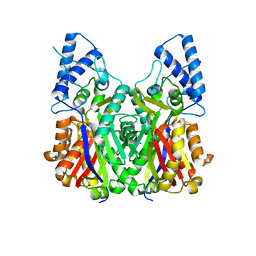 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3VS9
 
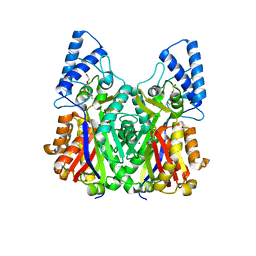 | | Crystal structure of type III PKS ArsC mutant | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3VGM
 
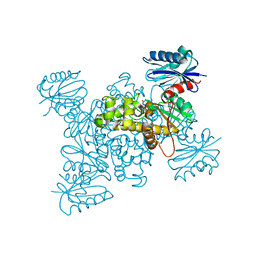 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, ZINC ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
3VGL
 
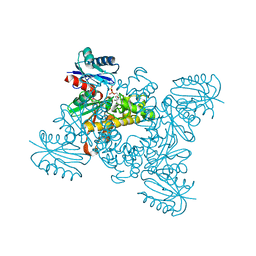 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus in complex with glucose and AMPPNP | | Descriptor: | Glucokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
3VGK
 
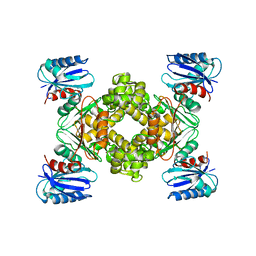 | | Crystal structure of a ROK family glucokinase from Streptomyces griseus | | Descriptor: | Glucokinase, SULFATE ION, ZINC ION | | Authors: | Miyazono, K, Tabei, N, Morita, S, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus
J.Bacteriol., 194, 2012
|
|
