4TMC
 
 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | 分子名称: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | 著者 | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | 登録日 | 2014-05-31 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4TMB
 
 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme | | 著者 | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | 登録日 | 2014-05-31 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
7F3A
 
 | | Arabidopsis thaliana GH1 beta-glucosidase AtBGlu42 | | 分子名称: | Beta-glucosidase 42, GLYCEROL | | 著者 | Horikoshi, S, Saburi, W, Yu, J, Yao, M. | | 登録日 | 2021-06-16 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Substrate specificity of glycoside hydrolase family 1 beta-glucosidase AtBGlu42 from Arabidopsis thaliana and its molecular mechanism.
Biosci.Biotechnol.Biochem., 86, 2022
|
|
2LDS
 
 | |
4NHX
 
 | | Crystal structure of human OGFOD1, 2-oxoglutarate and iron-dependent oxygenase domain containing 1, in complex with N-oxalylglycine (NOG) | | 分子名称: | 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Horita, S, McDonough, M.A, Schofield, C.J. | | 登録日 | 2013-11-05 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
2KSW
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for Oryctin | | 分子名称: | Oryctin | | 著者 | Horita, S, Ishibashi, J, Nagata, K, Miyakawa, T, Yamakawa, M, Tanokura, M. | | 登録日 | 2010-01-14 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Isolation, cDNA cloning, and structure-based functional characterization of oryctin, a hemolymph protein from the coconut rhinoceros beetle, Oryctes rhinoceros, as a novel serine protease inhibitor
J.Biol.Chem., 285, 2010
|
|
4NHY
 
 | | Crystal structure of human OGFOD1, 2-oxoglutarate and iron-dependent oxygenase domain containing 1, in complex with pyridine-2,4-dicarboxylic acid (2,4-PDCA) | | 分子名称: | 2-oxoglutarate and iron-dependent oxygenase domain-containing protein 1, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Horita, S, McDonough, M.A, Schofield, C.J. | | 登録日 | 2013-11-05 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Structure of the Ribosomal Oxygenase OGFOD1 Provides Insights into the Regio- and Stereoselectivity of Prolyl Hydroxylases.
Structure, 23, 2015
|
|
5B8C
 
 | | High resolution structure of the human PD-1 in complex with pembrolizumab Fv | | 分子名称: | Pembrolizumab heavy chain variable region (PemVH), Pembrolizumab light chain variable region (PemVL), Programmed cell death protein 1 | | 著者 | Horita, S, Shimamura, T, Iwata, S, Nomura, N. | | 登録日 | 2016-06-14 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.146 Å) | | 主引用文献 | High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1
Sci Rep, 6, 2016
|
|
6AGZ
 
 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | 著者 | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | 登録日 | 2018-08-15 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
3VYH
 
 | | Crystal structure of aW116R mutant of nitrile hydratase from Pseudonocardia thermophilla | | 分子名称: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit alpha, Cobalt-containing nitrile hydratase subunit beta | | 著者 | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | 登録日 | 2012-09-25 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
3VYG
 
 | | Crystal structure of Thiocyanate hydrolase mutant R136W | | 分子名称: | COBALT (III) ION, L(+)-TARTARIC ACID, Thiocyanate hydrolase subunit alpha, ... | | 著者 | Yamanaka, Y, Sato, M, Arakawa, T, Namima, S, Hori, S, Ohtaki, A, Noguchi, K, Katayama, Y, Yohda, M, Odaka, M. | | 登録日 | 2012-09-25 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Effects of argnine residue around the substrate pocket on the substrate specificity of thiocyanate hydrolase
To be published
|
|
5WS3
 
 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | 分子名称: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | 著者 | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | 登録日 | 2016-12-05 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | 分子名称: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | 著者 | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | 登録日 | 2016-11-25 | | 公開日 | 2017-11-29 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
4IJ5
 
 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase from Hydrogenobacter thermophilus TK-6 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoserine phosphatase 1 | | 著者 | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | 登録日 | 2012-12-21 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
4IJ6
 
 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase Mutant (H9A) from Hydrogenobacter thermophilus TK-6 in Complex with L-phosphoserine | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOSERINE, ... | | 著者 | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | 登録日 | 2012-12-21 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
3OIT
 
 | | Crystal structure of curcuminoid synthase CUS from Oryza sativa | | 分子名称: | Os07g0271500 protein | | 著者 | Miyazono, K, Um, J, Imai, F.L, Katsuyama, Y, Ohnishi, Y, Horinouchi, S, Tanokura, M. | | 登録日 | 2010-08-19 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of curcuminoid synthase CUS from Oryza sativa
Proteins, 79, 2011
|
|
7SD4
 
 | | SARS-CoV-2 Nucleocapsid N-terminal domain (N-NTD) protein | | 分子名称: | Nucleoprotein | | 著者 | Sarkar, S, Runge, B, Russell, R.W, Calero, D, Zeinalilathori, S, Quinn, C.M, Lu, M, Calero, G, Gronenborn, A.M, Polenova, T. | | 登録日 | 2021-09-29 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Atomic-Resolution Structure of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain.
J.Am.Chem.Soc., 144, 2022
|
|
3OV3
 
 | | G211F mutant of curcumin synthase 1 from Curcuma longa | | 分子名称: | Curcumin synthase, MALONATE ION | | 著者 | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2010-09-15 | | 公開日 | 2010-12-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
3OV2
 
 | | Curcumin synthase 1 from Curcuma longa | | 分子名称: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | 著者 | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2010-09-15 | | 公開日 | 2010-12-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
6KZA
 
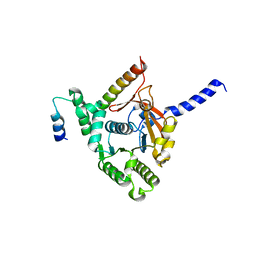 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | 分子名称: | DNA replication protein DnaC, Replicative DNA helicase | | 著者 | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | 登録日 | 2019-09-23 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
1UI6
 
 | |
1UI5
 
 | |
1ET8
 
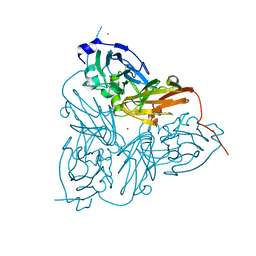 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASN MUTANT FROM ALCALIGENES FAECALIS | | 分子名称: | COPPER (II) ION, NITRITE REDUCTASE, ZINC ION | | 著者 | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | 登録日 | 2000-04-12 | | 公開日 | 2000-08-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
3A4I
 
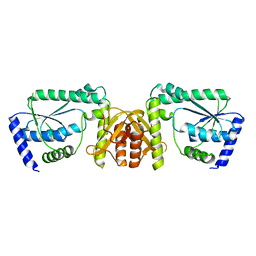 | | Crystal structure of GMP synthetase PH1347 from Pyrococcus horikoshii OT3 | | 分子名称: | GMP synthase [glutamine-hydrolyzing] subunit B | | 著者 | Maruoka, S, Horita, S, Lee, W.C, Nagata, K, Tanokura, M. | | 登録日 | 2009-07-07 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structure of the ATPPase subunit and its substrate-dependent association with the GATase Subunit: a novel regulatory mechanism for a two-subunit-type GMP synthetase from Pyrococcus horikoshii OT3.
J.Mol.Biol., 395, 2010
|
|
1ET7
 
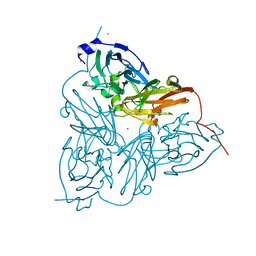 | | CRYSTAL STRUCTURE OF NITRITE REDUCTASE HIS255ASP MUTANT FROM ALCALIGENES FAECALIS S-6 | | 分子名称: | CADMIUM ION, COPPER (II) ION, NITRITE REDUCTASE | | 著者 | Boulanger, M.J, Kukimoto, M, Nishiyama, M, Horinouchi, S, Murphy, M.E.P. | | 登録日 | 2000-04-12 | | 公開日 | 2000-08-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase.
J.Biol.Chem., 275, 2000
|
|
