2X9I
 
 | |
2YJ6
 
 | |
2YBV
 
 | | STRUCTURE OF RUBISCO FROM THERMOSYNECHOCOCCUS ELONGATUS | | Descriptor: | CHLORIDE ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL SUBUNIT | | Authors: | Terlecka, B, Wilhelmi, V, Bialek, W, Gubernator, B, Szczepaniak, A, Hofmann, E. | | Deposit date: | 2011-03-10 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Ribulose-1,5-Bisphosphate Carboxylase Oxygenase from Thermosynechococcus Elongatus
To be Published
|
|
2WJK
 
 | | Bacteriorhodopsin mutant E204D | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Potschies, M, Hofmann, E, Gerwert, K. | | Deposit date: | 2009-05-27 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Directional proton transfer in membrane proteins achieved through protonated protein-bound water molecules: a proton diode.
Angew. Chem. Int. Ed. Engl., 49, 2010
|
|
2X21
 
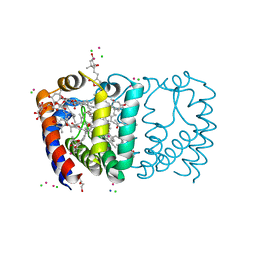 | | Structure of Peridinin-Chlorophyll-Protein reconstituted with BChl-a | | Descriptor: | BACTERIOCHLOROPHYLL A, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Schulte, T, Hiller, R.G, Hofmann, E. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-Ray Structures of the Peridinin-Chlorophyll-Protein Reconstituted with Different Chlorophylls.
FEBS Lett., 584, 2010
|
|
2X9O
 
 | | STRUCTURE OF 15, 16- DIHYDROBILIVERDIN:FERREDOXIN OXIDOREDUCTASE (PebA) | | Descriptor: | 15,16-DIHYDROBILIVERDIN-FERREDOXIN OXIDOREDUCTASE, BILIVERDINE IX ALPHA | | Authors: | Busch, A.W.U, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2010-03-23 | | Release date: | 2011-03-30 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Mechanistic Insight Into the Ferredoxin-Mediated Two-Electron Reduction of Bilins.
Biochem.J., 439, 2011
|
|
2X1Z
 
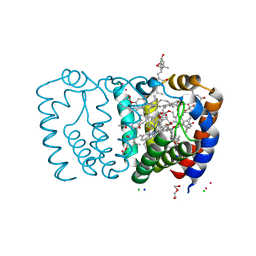 | | Structure of Peridinin-Chlorophyll-Protein reconstituted with Chl-d | | Descriptor: | CADMIUM ION, CHLORIDE ION, CHLOROPHYLL D, ... | | Authors: | Schulte, T, Hiller, R.G, Hofmann, E. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Peridinin-Chlorophyll-Protein Reconstituted with Different Chlorophylls.
FEBS Lett., 584, 2010
|
|
2X20
 
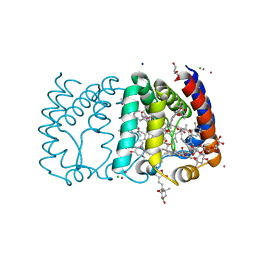 | | Structure of Peridinin-Chlorophyll-Protein reconstituted with Chl-b | | Descriptor: | CADMIUM ION, CHLORIDE ION, CHLOROPHYLL B, ... | | Authors: | Schulte, T, Hiller, R.G, Hofmann, E. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-Ray Structures of the Peridinin-Chlorophyll-Protein Reconstituted with Different Chlorophylls.
FEBS Lett., 584, 2010
|
|
2YJ4
 
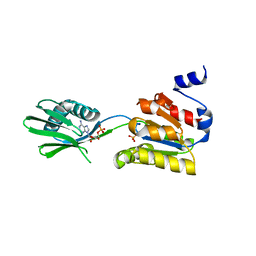 | | Conformational changes in the catalytic domain of the CPx-ATPase CopB- B upon nucleotide binding | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, COPPER-TRANSPORTING ATPASE, ... | | Authors: | Voellmecke, C, Schlicker, C, Luebben, M, Hofmann, E. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes in the Catalytic Domain of the Cpx-ATPase Copb-B Upon Nucleotide Binding
To be Published
|
|
2YJ5
 
 | |
2YJ3
 
 | | Conformational changes in the catalytic domain of the CPx-ATPase CopB- B upon nucleotide binding | | Descriptor: | COPPER-TRANSPORTING ATPASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Voellmecke, C, Schlicker, C, Luebben, M, Hofmann, E. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes in the Catalytic Domain of the Cpx-ATPase Copb-B Upon Nucleotide Binding
To be Published
|
|
