6ELF
 
 | | Tryptophan Repressor TrpR from E.coli variant M42F T44L T81I S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
6FAL
 
 | |
6F7F
 
 | |
6F7G
 
 | |
6F9K
 
 | |
2XH2
 
 | |
2XH0
 
 | |
2XH7
 
 | |
7OYZ
 
 | | E.coli's putrescine receptor variant PotF/D in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putrescine-binding periplasmic protein PotF, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYW
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88L S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYU
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S F88Y in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putrescine-binding periplasmic protein PotF, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYS
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
2XGZ
 
 | |
7OYX
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S F88Y S247D in complex with spermidine | | Descriptor: | (2~{R})-1-(2-methoxyethoxy)propan-2-amine, (2~{R})-1-[(2~{R})-1-[(2~{S})-1-[(2~{S})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-yl]oxypropan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYY
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutation S247D in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kroeger, P, Shanmugaratnam, S, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYT
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88L in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYV
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88A S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-yl]oxypropan-2-amine, (2~{S})-1-methoxypropan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
2XH4
 
 | |
7OT8
 
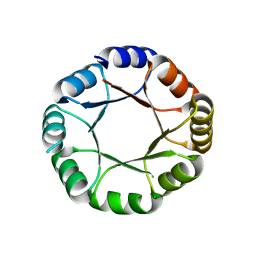 | |
7OSU
 
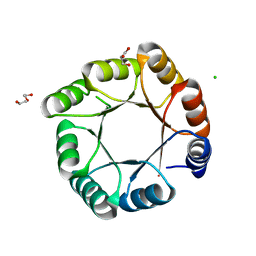 | | sTIM11noCys-SB, a de novo designed TIM barrel with a salt-bridge cluster (crystal form 1) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Romero-Romero, S, Kordes, S, Hocker, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
7OSV
 
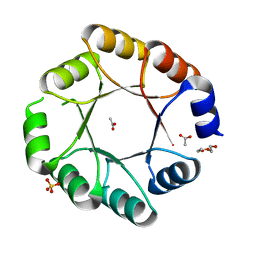 | | DeNovoTIM6-SB, a de novo designed TIM barrel with a salt-bridge cluster (crystal form 1) | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, DeNovoTIM6-SB, ... | | Authors: | Kordes, S, Romero-Romero, S, Hocker, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
7OT7
 
 | |
7P12
 
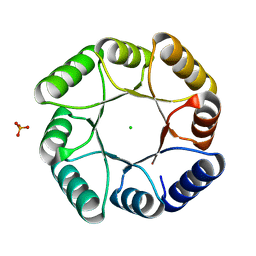 | | DeNovoTIM13-SB, a de novo designed TIM barrel with a salt-bridge cluster | | Descriptor: | CHLORIDE ION, DeNovoTIM13-SB, PHOSPHATE ION | | Authors: | Kordes, S, Romero-Romero, S, Hocker, B. | | Deposit date: | 2021-07-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
