1H2A
 
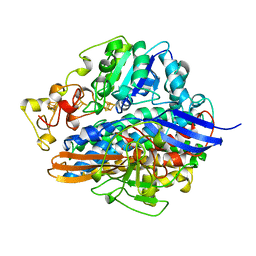 | | SINGLE CRYSTALS OF HYDROGENASE FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FE3-S4 CLUSTER, HYDROGENASE, IRON/SULFUR CLUSTER, ... | | Authors: | Higuchi, Y, Yasuoka, N. | | Deposit date: | 1997-10-17 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis.
Structure, 5, 1997
|
|
2RDV
 
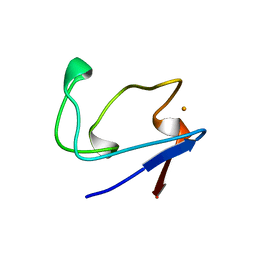 | |
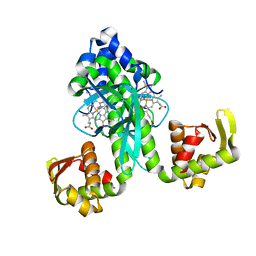
2FMY
 
 | |
1H2R
 
 | |
1RDV
 
 | |
1VEK
 
 | | Solution Structure of RSGI RUH-011, a UBA Domain from Arabidopsis cDNA | | Descriptor: | ubiquitin-specific protease 14, putative | | Authors: | Higuchi, Y, Abe, T, Hirota, H, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-31 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-011, a UBA Domain from Arabidopsis cDNA
To be Published
|
|
2CDV
 
 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | Deposit date: | 1983-11-15 | | Release date: | 1984-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
1WIV
 
 | | solution structure of RSGI RUH-023, a UBA domain from Arabidopsis cDNA | | Descriptor: | ubiquitin-specific protease 14 | | Authors: | Higuchi, Y, Abe, T, Hirota, H, Izumi, K, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | solution structure of RSGI RUH-023, a UBA domain from Arabidopsis cDNA
To be Published
|
|
1VEJ
 
 | | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA | | Descriptor: | RIKEN cDNA 4931431F19 | | Authors: | Higuchi, Y, Abe, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-31 | | Release date: | 2005-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA
To be Published
|
|
2EWI
 
 | |
2EWU
 
 | |
2FFN
 
 | |
2EWK
 
 | |
2YYW
 
 | |
2YYX
 
 | |
2Z47
 
 | |
2YXC
 
 | |
2YXW
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
2YXV
 
 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
7VXQ
 
 | | The Carbon Monoxide Complex of [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 | | Descriptor: | CARBON MONOXIDE, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Higuchi, K, Imanishi, T, Higuchi, Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and spectroscopic characterization of CO inhibition of [NiFe]-hydrogenase from Citrobacter sp. S-77.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6GHP
 
 | | 14-3-3sigma in complex with a TASK3 peptide stabilized by semi-synthetic natural product FC-NAc | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Potassium channel subfamily K member 9, ... | | Authors: | Andrei, S.A, de Vink, P.J, Brunsveld, L, Ottmann, C, Higuchi, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-01 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6L1V
 
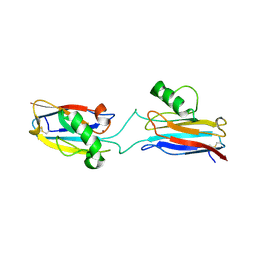 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | Descriptor: | Azurin-1, COPPER (II) ION | | Authors: | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
6LS8
 
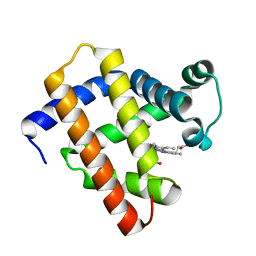 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTM
 
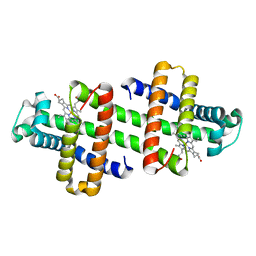 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTL
 
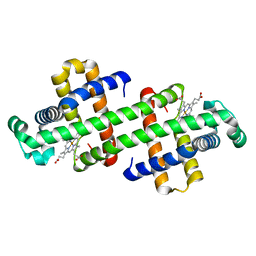 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
