8IBS
 
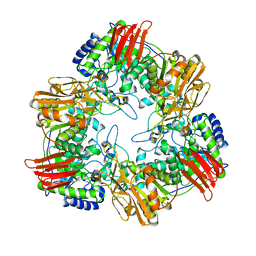 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E160A/E318A mutant in complex with galactose | | Descriptor: | Beta-galactosidase, alpha-D-galactopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBR
 
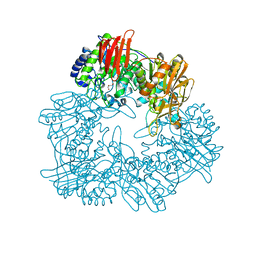 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis in complex with glycerol | | Descriptor: | Beta-galactosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
8IBT
 
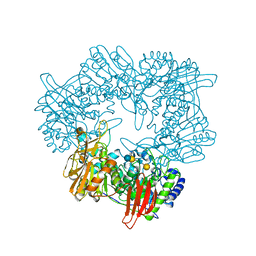 | | Crystal structure of GH42 beta-galactosidase BiBga42A from Bifidobacterium longum subspecies infantis E318S mutant in complex with lacto-N-tetraose | | Descriptor: | Beta-galactosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hidaka, M, Fushinobu, S, Gotoh, A, Katayama, T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mode of a glycoside hydrolase family 42 beta-galactosidase from Bifidobacterium longum subspecies infantis ( Bi Bga42A) revealed by crystallographic and mutational analyses.
Microbiome Res Rep, 2, 2023
|
|
3A3V
 
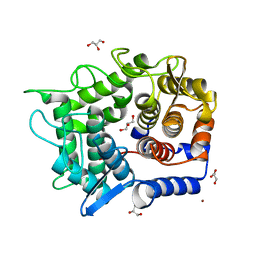 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
3ACT
 
 | |
3AFJ
 
 | |
3ACS
 
 | |
1V7V
 
 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase | | Descriptor: | CALCIUM ION, chitobiose phosphorylase | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
2CQS
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Ammonium Sulfate | | Descriptor: | Cellobiose Phosphorylase, SULFATE ION, beta-D-glucopyranose | | Authors: | Hidaka, M, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural dissection of the reaction mechanism of cellobiose phosphorylase.
Biochem.J., 398, 2006
|
|
2CQT
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate | | Descriptor: | Cellobiose Phosphorylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hidaka, M, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dissection of the reaction mechanism of cellobiose phosphorylase.
Biochem.J., 398, 2006
|
|
1V7W
 
 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7X
 
 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1KWK
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase in complex with galactose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trimeric crystal structure of the glycoside hydrolase family 42 beta-galactosidase from Thermus thermophilus A4 and the structure of its complex with galactose.
J.Mol.Biol., 322, 2002
|
|
1KWG
 
 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
2ZUT
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUW
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUV
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc, Ethylene glycol, and nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUS
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase | | Descriptor: | Lacto-N-biose phosphorylase, MAGNESIUM ION | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUU
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
3UES
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UET
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
8XW5
 
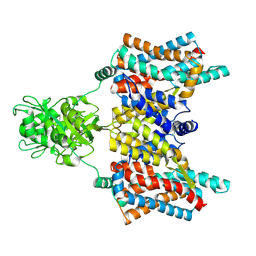 | | Cryo-EM structure of the aspartate:alanine antiporter AspT mutant L60C | | Descriptor: | Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
8Y1X
 
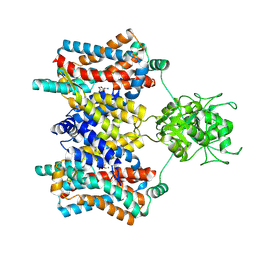 | | Cryo-EM structure of the aspartate:alanine antiporter AspT WT | | Descriptor: | ASPARTIC ACID, Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
1WU5
 
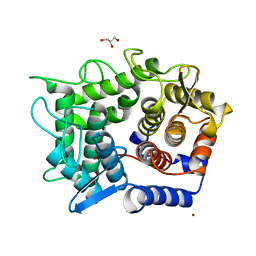 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase complexed with xylose | | Descriptor: | GLYCEROL, NICKEL (II) ION, beta-D-xylopyranose, ... | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
1WU4
 
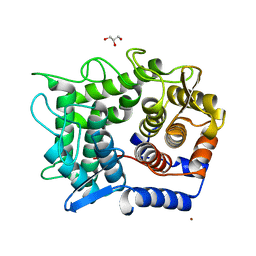 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase | | Descriptor: | GLYCEROL, NICKEL (II) ION, xylanase Y | | Authors: | Fushinobu, S, Hidaka, M, Honda, Y, Wakagi, T, Shoun, H, Kitaoka, M. | | Deposit date: | 2004-12-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for the Specificity of the Reducing End Xylose-releasing Exo-oligoxylanase from Bacillus halodurans C-125
J.Biol.Chem., 280, 2005
|
|
