3ZH3
 
 | |
3ZG5
 
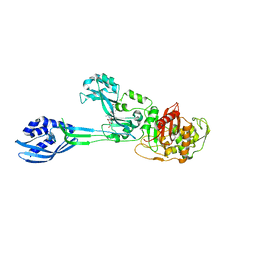 | | Crystal structure of PBP2a from MRSA in complex with peptidoglycan analogue at allosteric | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDOGLYCAN ANALOGUE, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ZHG
 
 | |
6FCU
 
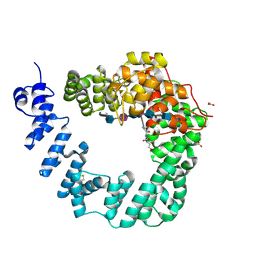 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5G2M
 
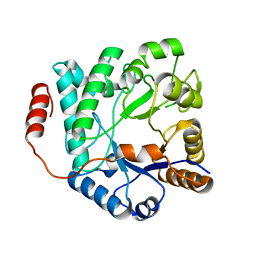 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5G5K
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-05-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
6FC4
 
 | |
6FCQ
 
 | |
6FCR
 
 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMtetrapeptide-NAG-anhNAMtetrapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FCS
 
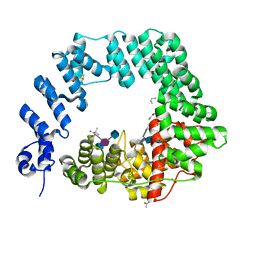 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMpentapeptide-NAG-NAMpentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FBT
 
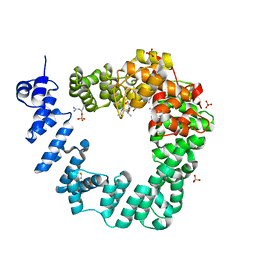 | | The X-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa in complex with the reaction product NAG-anhNAMpentapeptide | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, NAG-anhNAMpentapeptide, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ATF
 
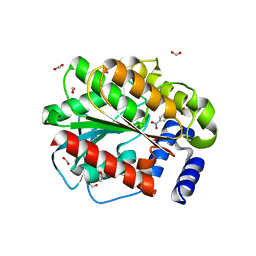 | | Structure of EstD11 in complex with p-Nitrophenol | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AV5
 
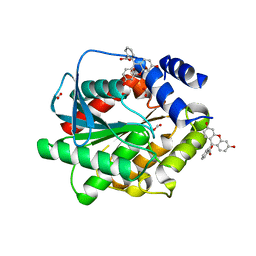 | | Structure of EstD11 in complex with Fluorescein | | Descriptor: | ACETATE ION, EstD11, FLUORESCIN, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT3
 
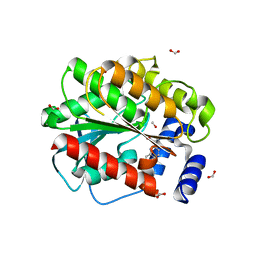 | | Structure of EstD11 in complex with Naproxen and methanol | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT0
 
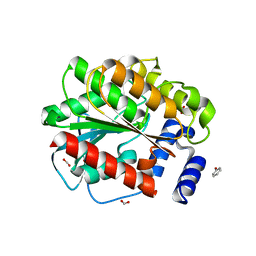 | |
7ATD
 
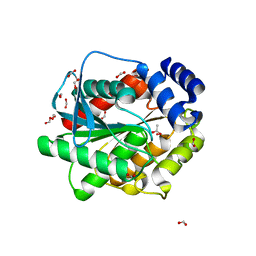 | | Structure of inactive EstD11 S144A in complex with methyl-naproxen | | Descriptor: | ACETATE ION, EstD11 S144A, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-29 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7ATQ
 
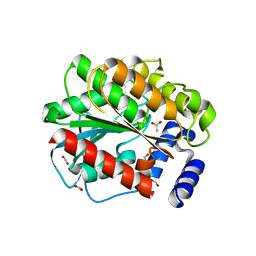 | | Structure of EstD11 in complex with cyclohexane carboxylic acid | | Descriptor: | ACETATE ION, EstD11, FORMIC ACID, ... | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-30 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AT4
 
 | | Structure of EstD11 in complex with Naproxen | | Descriptor: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, EstD11, FORMIC ACID | | Authors: | Miguel-Ruano, V, Rivera, I, Hermoso, J.A. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of a novel thermophilic esterase EstD11 provide catalytic insights for the HSL family.
Comput Struct Biotechnol J, 19, 2021
|
|
7AUY
 
 | |
7AT2
 
 | |
7AJZ
 
 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide) | | Descriptor: | 1,2-ETHANEDIOL, L,D-transpeptidase YcbB, NAG-NAM(tetrapeptide), ... | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide)
To Be Published
|
|
7AGZ
 
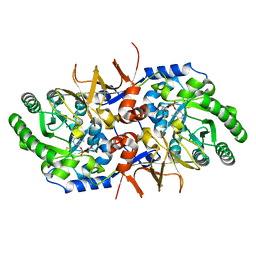 | | BsrV no-histagged | | Descriptor: | Broad specificity amino-acid racemase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Espaillat, A, Cava, F, Hermoso, J.A. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Binding of non-canonical peptidoglycan controls Vibrio cholerae broad spectrum racemase activity.
Comput Struct Biotechnol J, 19, 2021
|
|
7AJO
 
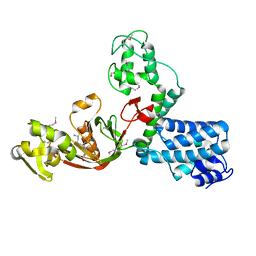 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with the cross-linking reaction intermediate | | Descriptor: | (2~{S},6~{S})-2-azanyl-6-[[(4~{R})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]heptanedioic acid, 1,2-ETHANEDIOL, L,D-transpeptidase YcbB | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with the cross-linking reaction intermediate
To Be Published
|
|
7AJX
 
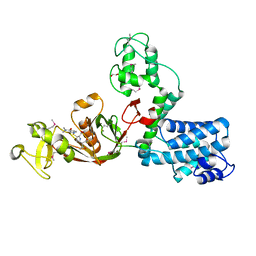 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, L,D-transpeptidase YcbB | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with meropenem
To Be Published
|
|
7AJ9
 
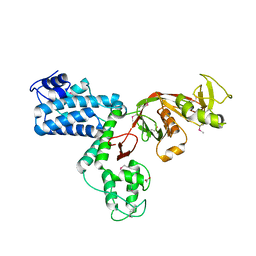 | |
