7X8C
 
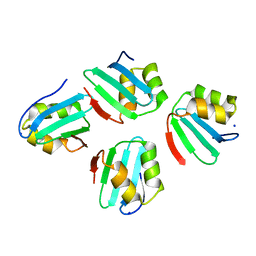 | | Crystal structure of a KTSC family protein from Euryarchaeon Methanolobus vulcani | | Descriptor: | KTSC domain-containing protein, SODIUM ION | | Authors: | Zhang, Z.F, Zhu, K.L, Chen, Y.Y, Cao, P, Gong, Y. | | Deposit date: | 2022-03-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Biochemical and structural characterization of a KTSC family single-stranded DNA-binding protein from Euryarchaea.
Int.J.Biol.Macromol., 216, 2022
|
|
7XI6
 
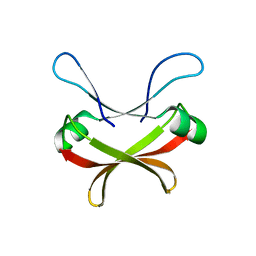 | | Crystal structure of C56 from pSSVi | | Descriptor: | C56 | | Authors: | Zhang, Z.F, Ren, Y, Chen, Y.Y, Zhang, X.W, Dong, Y.H, Gong, Y, Cao, P, Huang, L. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the AbrB-like protein C56 conserved in a novel family of integrated genetic elements in Sulfolobales
To Be Published
|
|
5WVZ
 
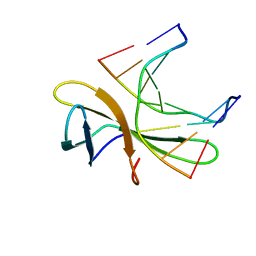 | | The crystal structure of Cren7 mutant L28F in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-30 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVY
 
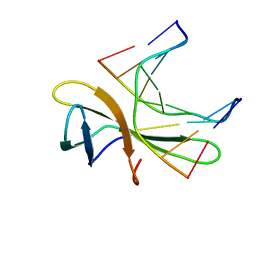 | | The crystal structure of Cren7 mutant L28V in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVW
 
 | | The crystal structure of Cren7 mutant L28A in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WWC
 
 | | The crystal structure of Cren7 mutant L28M in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-31 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
4R56
 
 | | Crystal structure of Sulfolobus Cren7-dsDNA(GTGATCAC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
4R55
 
 | | The crystal structure of a Cren7 mutant protein GR and dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Chen, Y.Y, Li, H.B, Huang, L. | | Deposit date: | 2014-08-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4
Extremophiles, 19, 2015
|
|
5K07
 
 | | Crystal structure of CREN7-DSDNA (GTAATTGC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea
PLoS ONE, 11, 2016
|
|
5K17
 
 | | Crystal structure of CREN7-DSDNA (GTGATCGC) complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sequence-Dependent T:G Base Pair Opening in DNA Double Helix Bound by Cren7, a Chromatin Protein Conserved among Crenarchaea
PLoS ONE, 11, 2016
|
|
3LWI
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
3LWH
 
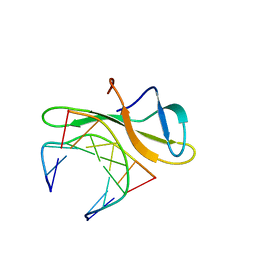 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
6A2H
 
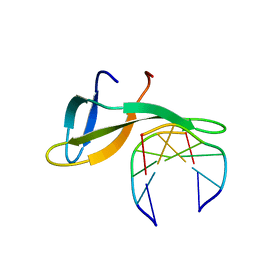 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(P*AP*AP*TP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*T)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
6A2I
 
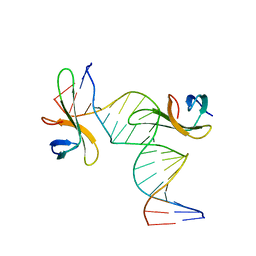 | | Architectural roles of Cren7 in folding crenarchaeal chromatin filament | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*CP*GP*TP*AP*GP*CP*TP*AP*AP*TP*TP*AP*GP*CP*TP*AP*CP*G)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Chen, Y.Y, Wang, L, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architectural roles of Cren7 in folding crenarchaeal chromatin filament.
Mol. Microbiol., 111, 2019
|
|
7W3T
 
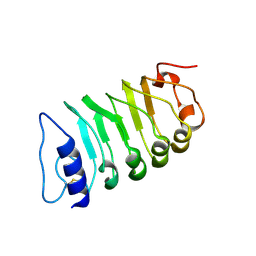 | | Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3X
 
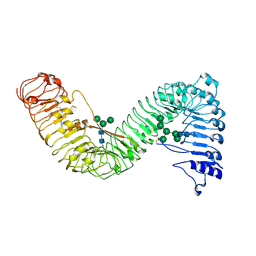 | | Cryo-EM structure of plant receptor like protein RXEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-localized LRR receptor-like protein, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7W3V
 
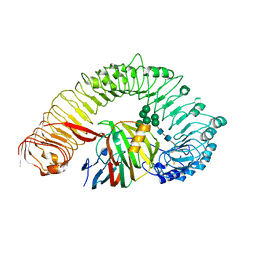 | | Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell 12A endoglucanase, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-22 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7DRB
 
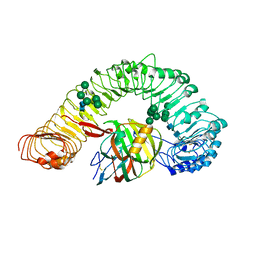 | | Crystal structure of plant receptor like protein RXEG1 with xyloglucanase XEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell 12A endoglucanase, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2020-12-27 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7DRC
 
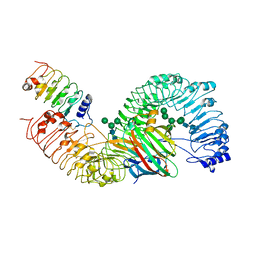 | | Cryo-EM structure of plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 and BAK1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2020-12-27 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7DFV
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7XDD
 
 | | Cryo-EM structure of EDS1 and PAD4 | | Descriptor: | Lipase-like PAD4, Protein EDS1 | | Authors: | Huang, S.J, Jia, A.L, Sun, Y, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-03-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
7XJP
 
 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7D3Y
 
 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
