7EPO
 
 | | Ketosteroid Isomerase KSI with 5-nitrobenzoxazole (5NBI) | | Descriptor: | 5-nitro-1,2-benzoxazole, SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
7EPN
 
 | | Ketosteroid Isomerase KSI native | | Descriptor: | SnoaL-like domain-containing protein | | Authors: | Liang, Y, Zhang, Q, Bartlam, M. | | Deposit date: | 2021-04-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization and Kemp eliminase activity of the Mycobacterium smegmatis Ketosteroid Isomerase.
Biochem.Biophys.Res.Commun., 560, 2021
|
|
2L1V
 
 | |
4QN1
 
 | | Crystal Structure of a Functionally Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH | | Descriptor: | E3 ubiquitin-protein ligase SHPRH, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zhang, Q, Li, Y, Walker, J.R, Guan, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a Function Uncharacterized Domain of E3 Ubiquitin Ligase SHPRH
To be Published
|
|
2JVA
 
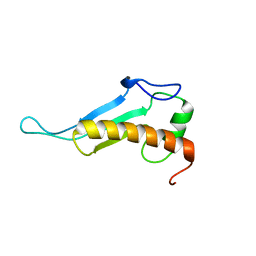 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | Descriptor: | Peptidyl-tRNA hydrolase domain protein | | Authors: | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
6IN9
 
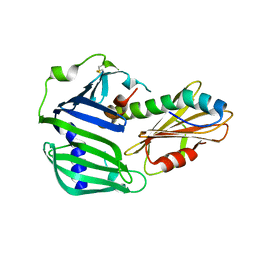 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6IN7
 
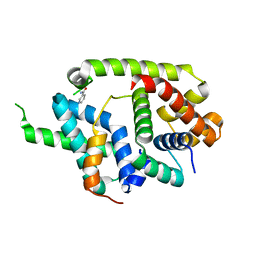 | | Crystal structure of AlgU in complex with MucA(cyto) | | Descriptor: | NICOTINAMIDE, RNA polymerase sigma-H factor, Sigma factor AlgU negative regulatory protein | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
7V4T
 
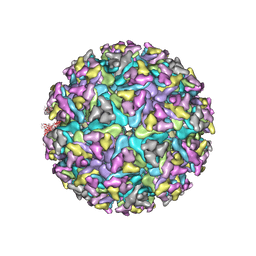 | | Cryo-EM structure of Alphavirus M1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Gao, Y, Jia, X, Zhang, Q. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Cryo-EM evidence of viral N-glycosylation reveal receptor binding mechanisms
of alphavirus M1
To Be Published
|
|
6IKK
 
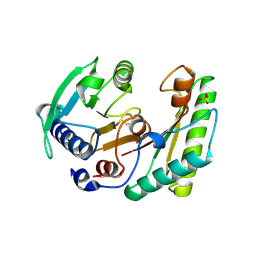 | |
6IKI
 
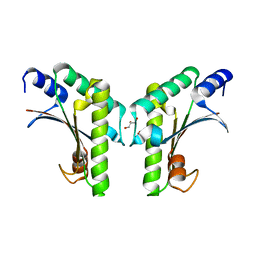 | | Crystal structure of YfiB(W55L) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IDL
 
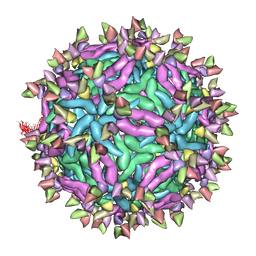 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class II particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
6IN8
 
 | | Crystal structure of MucB | | Descriptor: | Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6IDI
 
 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 8.0. | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
8GV3
 
 | | The cryo-EM structure of GSNOR with NYY001 | | Descriptor: | (4P)-4-{2-[4-(1H-imidazol-1-yl)phenyl]-5-[3-oxo-3-(2-oxo-1,3-thiazolidin-3-yl)propyl]-1H-pyrrol-1-yl}-3-methylbenzamide, Alcohol dehydrogenase class-3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Xia, Y, Zhang, Q, Yao, D, Zhao, S, Xie, L, Ji, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The cryo-EM structure of GSNOR with NYY001
To Be Published
|
|
6IDK
 
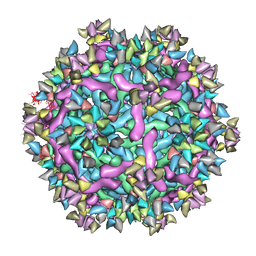 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class I particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
6IKJ
 
 | | Crystal structure of YfiB(F48S) | | Descriptor: | GLYCEROL, SULFATE ION, YfiB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis of activating mutants of YfiB from Pseudomonas aeruginosa PAO1.
Biochem. Biophys. Res. Commun., 506, 2018
|
|
5YKR
 
 | |
5YKT
 
 | |
4NYL
 
 | |
