8KFU
 
 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFV
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFR
 
 | | Crystal structure of ZmMOC1/nicked Holliday junction/Ca2+ complex | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (25-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFW
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 600 seconds | | 分子名称: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (33-MER), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFS
 
 | | Crystal structure of ZmMOC1/nicked Holliday junction complex at ground state | | 分子名称: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | 著者 | Zhang, D, Luo, Z, Lin, Z. | | 登録日 | 2023-08-16 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
5ISY
 
 | | Crystal structure of Nudix family protein with NAD | | 分子名称: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Zhang, D, Guan, Z, Zou, T, Yin, P. | | 登録日 | 2016-03-15 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
5UG1
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain with methylsulfonyl adduct | | 分子名称: | Acyltransferase, SODIUM ION, methanesulfonic acid | | 著者 | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | 登録日 | 2017-01-06 | | 公開日 | 2017-10-25 | | 最終更新日 | 2017-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UFY
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | 分子名称: | Acyltransferase, SODIUM ION | | 著者 | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | 登録日 | 2017-01-06 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
6CZV
 
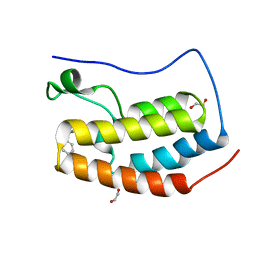 | | BRD4(BD1) complexed with 2759 | | 分子名称: | 1,2-ETHANEDIOL, 1-benzyl-5-(3,5-dimethyl-1,2-oxazol-4-yl)pyridin-2(1H)-one, Bromodomain-containing protein 4 | | 著者 | Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2018-04-09 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
8P3S
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | 著者 | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
6CZU
 
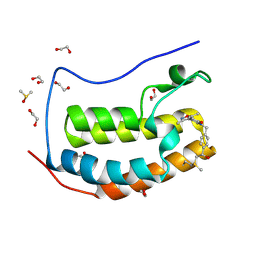 | | BRD4(BD1) complexed with 3219 | | 分子名称: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-({4-[(1R)-1-hydroxyethyl]phenyl}methyl)pyridin-2(1H)-one, Bromodomain-containing protein 4, ... | | 著者 | Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2018-04-09 | | 公開日 | 2018-09-26 | | 最終更新日 | 2018-10-10 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
8P3V
 
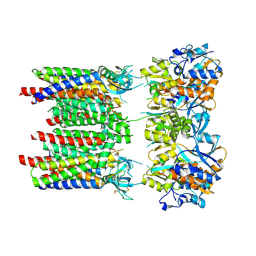 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 3 | | 分子名称: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | 著者 | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8PIV
 
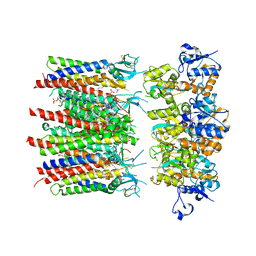 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor, ... | | 著者 | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | 登録日 | 2023-06-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3U
 
 | |
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | 分子名称: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | 著者 | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3W
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 4 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | 著者 | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Q
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | 著者 | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | 登録日 | 2023-05-18 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
1HTD
 
 | | STRUCTURAL INTERACTION OF NATURAL AND SYNTHETIC INHIBITORS WITH THE VENOM METALLOPROTEINASE, ATROLYSIN C (HT-D) | | 分子名称: | ATROLYSIN C, CALCIUM ION, ZINC ION | | 著者 | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | 登録日 | 1994-01-20 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1ATL
 
 | | Structural interaction of natural and synthetic inhibitors with the VENOM METALLOPROTEINASE, ATROLYSIN C (FORM-D) | | 分子名称: | CALCIUM ION, O-methyl-N-[(2S)-4-methyl-2-(sulfanylmethyl)pentanoyl]-L-tyrosine, Snake venom metalloproteinase atrolysin-D, ... | | 著者 | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | 登録日 | 1995-05-26 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1HYN
 
 | |
8SPM
 
 | | Crystal structure of NikA in complex Ni-AMA | | 分子名称: | Aspergillomarasmine A, NICKEL (II) ION, Nickel ABC transporter, ... | | 著者 | Sychantha, D, Prehna, G, Wright, G.D. | | 登録日 | 2023-05-03 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Targeting bacterial nickel transport with aspergillomarasmine A suppresses virulence-associated Ni-dependent enzymes.
Nat Commun, 15, 2024
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | 分子名称: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
8EKM
 
 | |
8EKK
 
 | |
