5WUQ
 
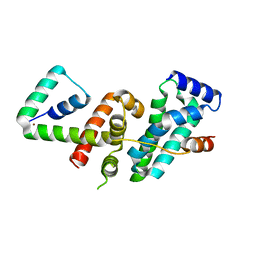 | | Crystal structure of SigW in complex with its anti-sigma RsiW, a zinc binding form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW, ZINC ION | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5WUR
 
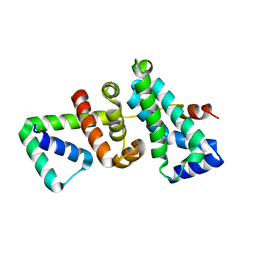 | | Crystal structure of SigW in complex with its anti-sigma RsiW, an oxdized form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
3WT4
 
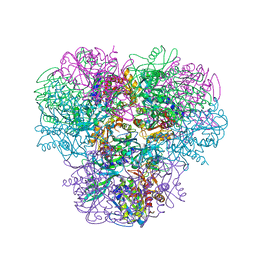 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
1PCV
 
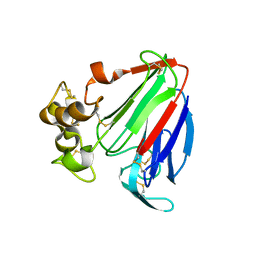 | | Crystal structure of osmotin, a plant antifungal protein | | Descriptor: | osmotin | | Authors: | Min, K, Ha, S.C, Yun, D.-J, Bressan, R.A, Kim, K.K. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of osmotin, a plant antifungal protein
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3R4Y
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3D1N
 
 | | Structure of human Brn-5 transcription factor in complex with corticotrophin-releasing hormone gene promoter | | Descriptor: | 5'-D(*DAP*DGP*DCP*DAP*DTP*DAP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DA)-3', 5'-D(*DTP*DTP*DAP*DTP*DTP*DAP*DTP*DTP*DTP*DAP*DTP*DGP*DCP*DT)-3', POU domain, ... | | Authors: | Pereira, J.H, Ha, S.C, Kim, S.-H. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human Brn-5 transcription factor in complex with CRH gene promoter.
J.Struct.Biol., 167, 2009
|
|
