4RYM
 
 | |
4RYJ
 
 | |
4RYO
 
 | | Crystal structure of BcTSPO type II high resolution monomer | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYR
 
 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom with DMSO | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYN
 
 | | Crystal structure of BcTSPO, type1 monomer | | Descriptor: | CACODYLATE ION, DODECYL-ALPHA-D-MALTOSIDE, Integral membrane protein, ... | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4XZC
 
 | | The crystal structure of Kupe virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZE
 
 | | The crystal structure of Hazara virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZA
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZ8
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
1SL4
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
8WNG
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile | | Descriptor: | ACETATE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO3
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Mupirocin | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNF
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in apo form | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNI
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO2
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNJ
 
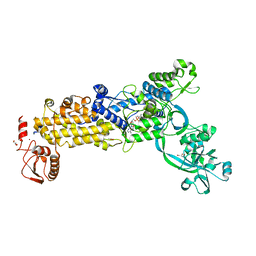 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
4RYI
 
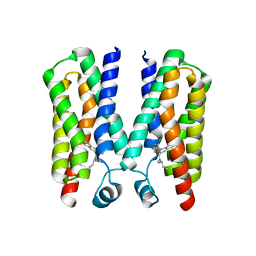 | |
4JYP
 
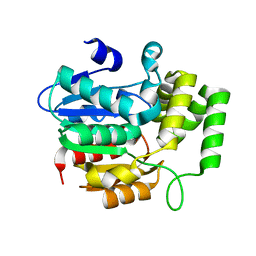 | | crystal Structure of KAI2 Apo form | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYM
 
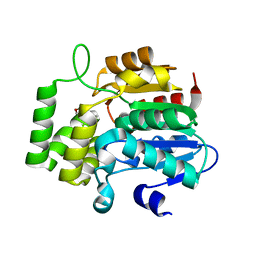 | | crystal Structure of KAI2 in complex with 3-methyl-2H-furo[2,3-c]pyran-2-one | | Descriptor: | 3-methyl-2H-furo[2,3-c]pyran-2-one, Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-30 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SL6
 
 | | Crystal Structure of a fragment of DC-SIGNR (containg the carbohydrate recognition domain and two repeats of the neck) complexed with Lewis-x. | | Descriptor: | C-type lectin DC-SIGNR, CALCIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
5TRL
 
 | |
5TRM
 
 | |
4TKR
 
 | | Native-SAD phasing for ThiT from Listeria monocytogenes serovar. | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, THIAMINE DIPHOSPHATE, Thiamine transporter thia | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
