1TAP
 
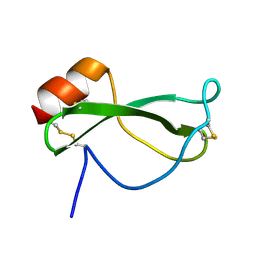 | | NMR SOLUTION STRUCTURE OF RECOMBINANT TICK ANTICOAGULANT PROTEIN (RTAP), A FACTOR XA INHIBITOR FROM THE TICK ORNITHODOROS MOUBATA | | Descriptor: | FACTOR XA INHIBITOR | | Authors: | Antuch, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1994-08-16 | | Release date: | 1994-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the recombinant tick anticoagulant protein (rTAP), a factor Xa inhibitor from the tick Ornithodoros moubata.
FEBS Lett., 352, 1994
|
|
1SZL
 
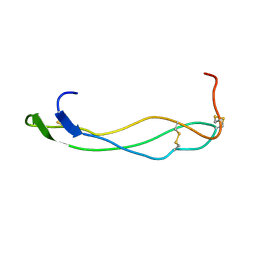 | | F-spondin TSR domain 1 | | Descriptor: | F-spondin | | Authors: | Tossavainen, H, Paakkonen, K, Permi, P, Kilpelainen, I, Guntert, P. | | Deposit date: | 2004-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and fourth TSR domains of F-spondin
Proteins, 64, 2006
|
|
1VEX
 
 | | F-spondin TSR domain 4 | | Descriptor: | F-spondin | | Authors: | Paakkonen, K, Tossavainen, H, Permi, P, Kilpelainen, I, Guntert, P. | | Deposit date: | 2004-04-06 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and fourth TSR domains of F-spondin
Proteins, 64, 2006
|
|
1PIT
 
 | |
1K91
 
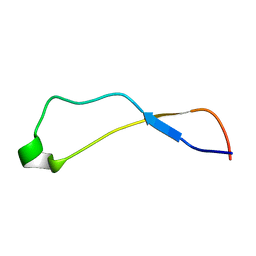 | | Solution Structure of Calreticulin P-domain subdomain (residues 221-256) | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Bettendorff, P, Braun, D, Herrmann, T, Fiorito, F, Guntert, P, Helenius, A, Wuthrich, K. | | Deposit date: | 2001-10-26 | | Release date: | 2002-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of 36 and 73-residue Fragments of the Calreticulin P-domain
J.Mol.Biol., 322, 2002
|
|
1K9C
 
 | | Solution Structure of Calreticulin P-domain subdomain (residues 189-261) | | Descriptor: | CALRETICULIN | | Authors: | Ellgaard, L, Bettendorff, P, Braun, D, Herrmann, T, Fiorito, F, Guntert, P, Helenius, A, Wuthrich, K. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of 36 and 73-residue Fragments of the Calreticulin P-domain
J.Mol.Biol., 322, 2002
|
|
1SR3
 
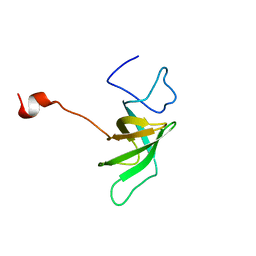 | |
2RQO
 
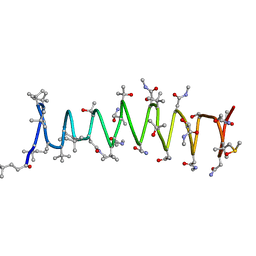 | | Solution structure of Polytheonamide B | | Descriptor: | polytheonamide B | | Authors: | Hamada, N, Matsunaga, S, Fujiwara, M, Fujjita, K, Hirota, H, Schmucki, R, Guntert, P, Fusetani, N. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Polytheonamide B, a Highly Cytotoxic Nonribosomal Polypeptide from Marine Sponge
J.Am.Chem.Soc., 2010
|
|
1BF8
 
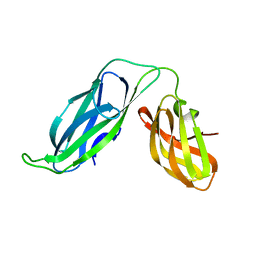 | | PERIPLASMIC CHAPERONE FIMC, NMR, 20 STRUCTURES | | Descriptor: | CHAPERONE PROTEIN FIMC | | Authors: | Pellecchia, M, Guntert, P, Glockshuber, R, Wuthrich, K. | | Deposit date: | 1998-05-28 | | Release date: | 1998-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the periplasmic chaperone FimC.
Nat.Struct.Biol., 5, 1998
|
|
5LAM
 
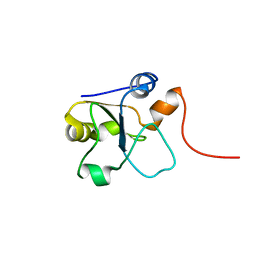 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAO
 
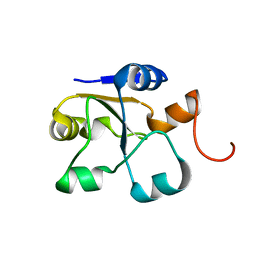 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
1Q9G
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX
J.Mol.Biol., 336, 2004
|
|
1WQU
 
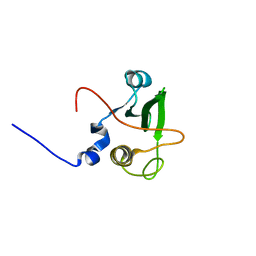 | | Solution structure of the human FES SH2 domain | | Descriptor: | Proto-oncogene tyrosine-protein kinase FES/FPS | | Authors: | Scott, A, Pantoja-Uceda, D, Koshiba, S, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Tanaka, A, Sugano, S, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-02 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Src homology 2 domain from the human feline sarcoma oncogene Fes
J.Biomol.NMR, 31, 2005
|
|
1QJK
 
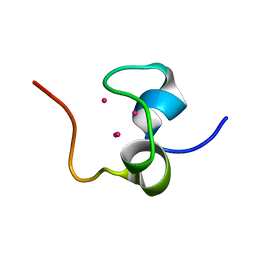 | | Metallothionein MTA from sea urchin (alpha domain) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
1QJL
 
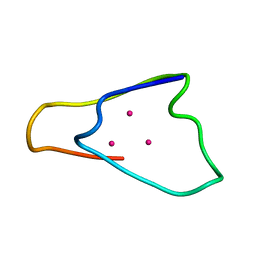 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | Descriptor: | CADMIUM ION, METALLOTHIONEIN | | Authors: | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
2RSM
 
 | | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65 (ICT2) | | Descriptor: | Probable peptide chain release factor C12orf65 homolog, mitochondrial | | Authors: | Enomoto, M, Tochio, N, Tomizawa, T, Koshiba, S, Guntert, P, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65.
Proteins, 2012
|
|
1E1W
 
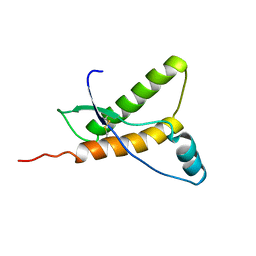 | | Human prion protein variant R220K | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-11 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E1U
 
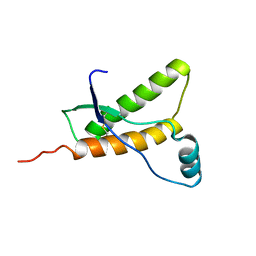 | | Human prion protein variant R220K | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-11 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E1G
 
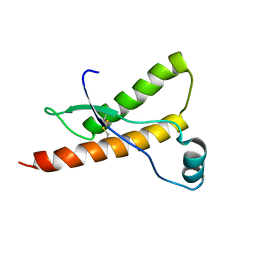 | | Human prion protein variant M166V | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-04 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E1J
 
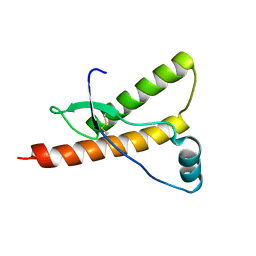 | | Human prion protein variant M166V | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-09 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E1P
 
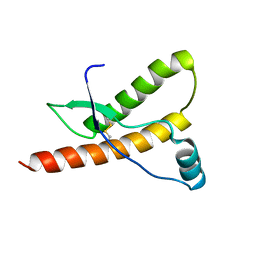 | | Human prion protein variant S170N | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-09 | | Release date: | 2000-07-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E1S
 
 | | Human prion protein variant S170N | | Descriptor: | PRION PROTEIN | | Authors: | Calzolai, L, Lysek, D.A, Guntert, P, Von Schroetter, C, Zahn, R, Riek, R, Wuthrich, K. | | Deposit date: | 2000-05-10 | | Release date: | 2000-07-21 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Three Single-Residue Variants of the Human Prion Protein
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1WKT
 
 | |
6F98
 
 | | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1 | | Descriptor: | E3 ubiquitin-protein ligase, ZINC ION | | Authors: | Kniss, A, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RING domain of the E3 ubiquitin ligase HRD1
To Be Published
|
|
1Q9F
 
 | | NMR STRUCTURE OF THE OUTER MEMBRANE PROTEIN OMPX IN DHPC MICELLES | | Descriptor: | Outer membrane protein X | | Authors: | Fernandez, C, Hilty, C, Wider, G, Guntert, P, Wuthrich, K. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the integral membrane protein OmpX.
J.Mol.Biol., 336, 2004
|
|
