6V33
 
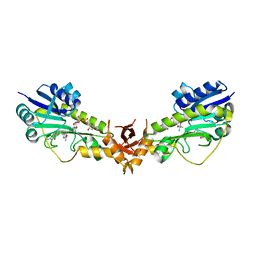 | | X-ray structure of a sugar N-formyltransferase from Pseudomonas congelans | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, dTDP-4-amino-4,6-dideoxyglucose, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6V2T
 
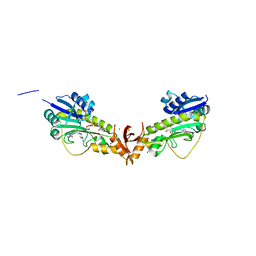 | | X-ray structure of a sugar N-formyltransferase from Shewanella sp FDAARGOS_354 | | Descriptor: | 1,2-ETHANEDIOL, FOLIC ACID, PHOSPHATE ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Misannotations of the genes encoding sugar N-formyltransferases.
Protein Sci., 29, 2020
|
|
6PZ2
 
 | |
7M15
 
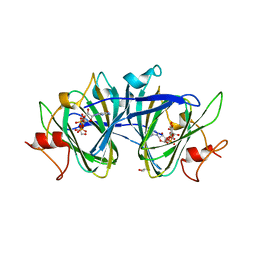 | | crystal structure of cj1430 in the presence of GDP-D-glycero-L-gluco-heptose, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GDP-D-glycero-L-gluco-heptose, [(2R,3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4R,5R,6S)-6-[(1R)-1,2-dihydroxyethyl]-3,4,5-trihydroxyoxan-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
7M14
 
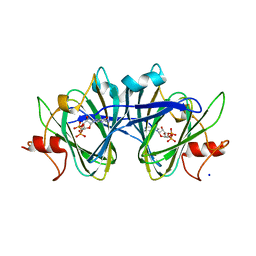 | | x-ray structure of cj1430 in the presence of GDP, a GDP-D-glycero-4-keto-D-lyxo-heptose-3,5-epimerase from campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Girardi, N.M, Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of d- glycero -l- gluco -Heptose in the Capsular Polysaccharides of Campylobacter jejuni .
Biochemistry, 60, 2021
|
|
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
2N3Q
 
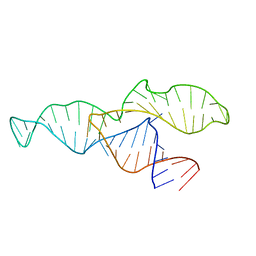 | |
2N3R
 
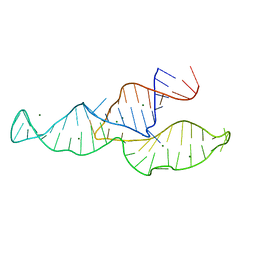 | | NMR structure of the II-III-VI three-way junction from the VS ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement | | Descriptor: | MAGNESIUM ION, RNA (62-MER) | | Authors: | Bonneau, E, Girard, N, Lemieux, S, Legault, P. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure.
Rna, 21, 2015
|
|
