5A9C
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra base domain deleted mutant | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A98
 
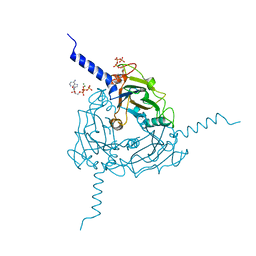 | | Crystal structure of Trichoplusia ni CPV15 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A96
 
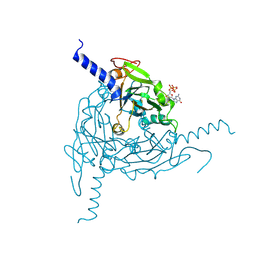 | | Crystal structure of Lymantria dispar CPV14 polyhedra | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9B
 
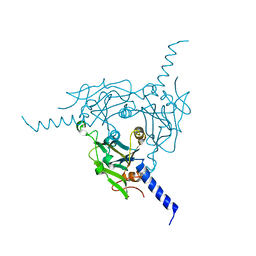 | | Crystal structure of Bombyx mori CPV1 polyhedra base domain deleted mutant | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9A
 
 | | Crystal structure of Simulium ubiquitum CPV20 polyhedra | | Descriptor: | POLYHEDRIN, URIDINE 5'-TRIPHOSPHATE | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8V
 
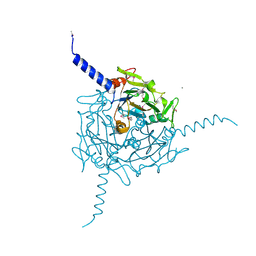 | | Crystal structure of Orgyia pseudotsugata CPV5 polyhedra with SeMet substitution | | Descriptor: | CALCIUM ION, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.074 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A9P
 
 | | Crystal structure of Operophtera brumata CPV18 polyhedra | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8T
 
 | | Crystal structure of Antheraea mylitta CPV4 polyhedra type 2 | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5A8U
 
 | | Crystal structure of Orgyia pseudotsugata CPV5 polyhedra | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Axford, D, Owen, R, Evans, G, Ginn, H.M, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | Polyhedra Structures and the Evolution of the Insect Viruses.
J.Struct.Biol., 192, 2015
|
|
5ACA
 
 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
7GTV
 
 | |
7GS8
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000466a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N},~{N},5,6-tetramethylthieno[2,3-d]pyrimidin-4-amine | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTH
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000637a | | Descriptor: | (6aR,8R,12R,12aS)-2-methyl-6a,10,11,12a-tetrahydro-6H,7H,9H-[1]benzopyrano[4,3-c]pyrazolo[1,2-a]pyrazol-9-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSD
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000605a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(5-amino-1,3,4-thiadiazol-2-yl)phenol, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GT0
 
 | |
7GTA
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000065a | | Descriptor: | (5S)-N-(4-fluorophenyl)-5-methyl-4,5-dihydro-1,3-thiazol-2-amine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTL
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000554a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, benzyl (3aS,8aS)-1-oxooctahydropyrrolo[3,4-d]azepine-6(1H)-carboxylate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSF
 
 | |
7GSZ
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMSOA000686b | | Descriptor: | 1-[4-methyl-2-(pyridin-4-yl)-1,3-thiazol-5-yl]methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSO
 
 | |
7GSW
 
 | |
7GU5
 
 | |
7GU4
 
 | |
7GTQ
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000311a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N}-(2-ethyl-1,2,3,4-tetrazol-5-yl)butanamide | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GU7
 
 | |
