6BTM
 
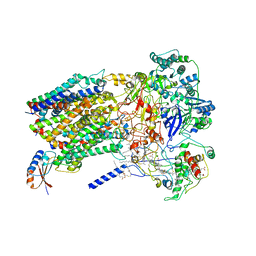 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | 著者 | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | 登録日 | 2017-12-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
5V33
 
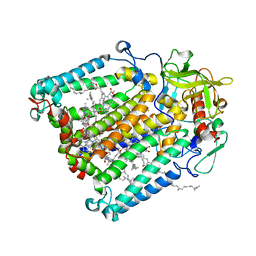 | | R. sphaeroides photosythetic reaction center mutant - Residue L223, Ser to Trp - Room Temperature Structure Solved on X-ray Transparent Microfluidic Chip | | 分子名称: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | 著者 | Schieferstein, J.M, Pawate, A.S, Sun, C, Wan, F, Broecker, J, Ernst, O.P, Gennis, R.B, Kenis, P.J.A. | | 登録日 | 2017-03-06 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.487 Å) | | 主引用文献 | X-ray transparent microfluidic chips for high-throughput screening and optimization of in meso membrane protein crystallization.
Biomicrofluidics, 11, 2017
|
|
7CUW
 
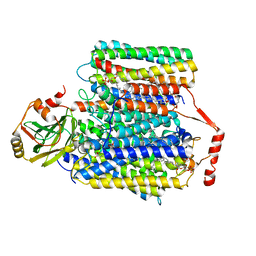 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-25 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUQ
 
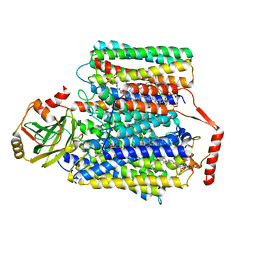 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-24 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
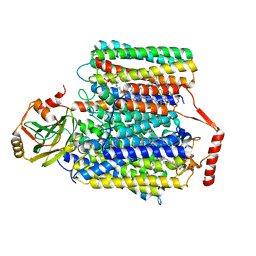 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-22 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2LTQ
 
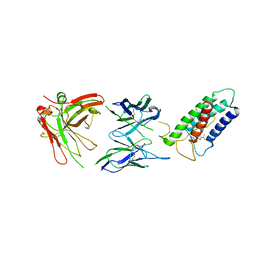 | | High resolution structure of DsbB C41S by joint calculation with solid-state NMR and X-ray data | | 分子名称: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | 著者 | Tang, M, Sperling, L.J, Schwieters, C.D, Nesbitt, A.E, Gennis, R.B, Rienstra, C.M. | | 登録日 | 2012-05-30 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer.
J.Mol.Biol., 425, 2013
|
|
2LEG
 
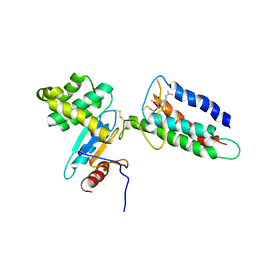 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | 分子名称: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | 著者 | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | 登録日 | 2011-06-15 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
6KOE
 
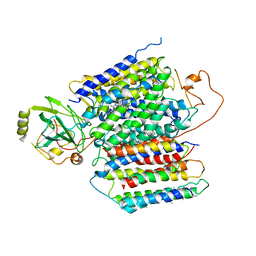 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | 分子名称: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | 著者 | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | 登録日 | 2019-08-09 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.75 Å) | | 主引用文献 | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOC
 
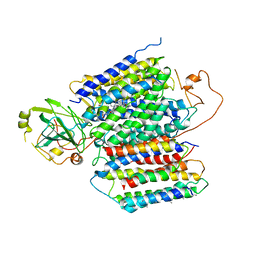 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | 分子名称: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | 著者 | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | 登録日 | 2019-08-09 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOB
 
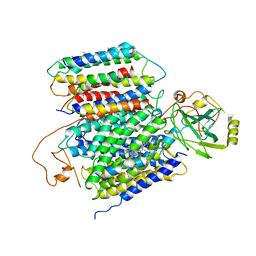 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | 分子名称: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | 著者 | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | 登録日 | 2019-08-09 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4O9U
 
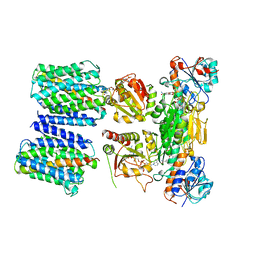 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | 分子名称: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | 著者 | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | 登録日 | 2014-01-02 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (6.926 Å) | | 主引用文献 | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4O9T
 
 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | 分子名称: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | 著者 | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | 登録日 | 2014-01-02 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.079 Å) | | 主引用文献 | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4O9P
 
 | | Crystal structure of Thermus thermophilis transhydrogeanse domain II dimer SeMet derivative | | 分子名称: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | 著者 | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | 登録日 | 2014-01-02 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4O93
 
 | | Crystal structure of Thermus thermophilis transhydrogeanse domain II dimer | | 分子名称: | MERCURY (II) ION, NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | 著者 | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | 登録日 | 2013-12-31 | | 公開日 | 2014-12-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4J1T
 
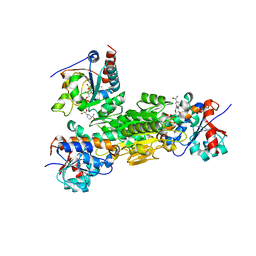 | | Crystal structure of Thermus thermophilus transhydrogenase heterotrimeric complex of the Alpha1 subunit dimer with the NADP binding domain (domain III) of the Beta subunit in P2(1) | | 分子名称: | GLYCEROL, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | 著者 | Yamaguchi, M, Leung, J, Schurig Briccio, L.A, Gennis, R.B, Stout, C.D. | | 登録日 | 2013-02-02 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal structure analysis of Thermus thermophilus transhydrogenase soluble domains
To be Published
|
|
4J16
 
 | | Crystal structure of Thermus thermophilus transhydrogenase heterotrimeric complex of the Alpha1 subunit dimer with the NADP binding domain (domain III) of the Beta subunit | | 分子名称: | CHLORIDE ION, GLYCEROL, NAD(P) transhydrogenase subunit beta, ... | | 著者 | Yamaguchi, M, Leung, J, Schurig Briccio, L.A, Gennis, R.B, Stout, C.D. | | 登録日 | 2013-02-01 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structure analysis of Thermus thermophilus transhydrogenase soluble domains
To be Published
|
|
