5YTD
 
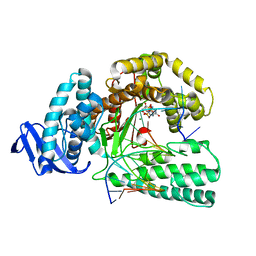 | | large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with the natural base pair 5fC:dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*(5FC)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTF
 
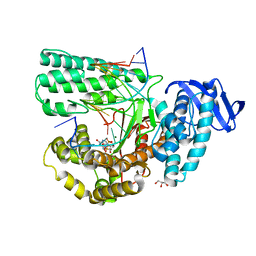 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTG
 
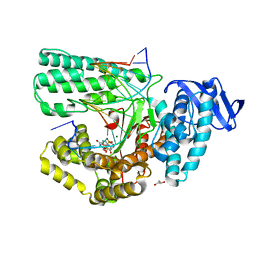 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5YTH
 
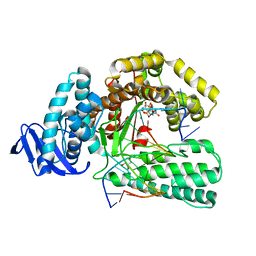 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base M-fC pair with dG | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(92F)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*GP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
8GZJ
 
 | |
8GZL
 
 | |
8GZM
 
 | |
8GZK
 
 | |
7DLZ
 
 | | Crystal Structure of Methyltransferase Ribozyme | | Descriptor: | RNA (45-MER), U1 small nuclear ribonucleoprotein A | | Authors: | Gan, J.H, Gao, Y.Q, Jiang, H.Y, Chen, D.R, Murchie, A.I.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7DE0
 
 | |
7DWH
 
 | | Complex structure of SAM-dependent methyltransferase ribozyme | | Descriptor: | COPPER (II) ION, RNA (45-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Jiang, H.Y, Gao, Y.Q, Chen, D.R, Murchie, A. | | Deposit date: | 2021-01-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7D32
 
 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D31
 
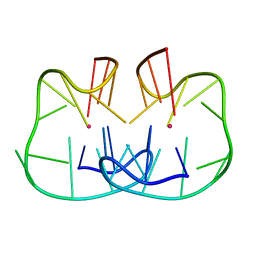 | | The TBA-Pb2+ complex in P41212 space group | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
7D33
 
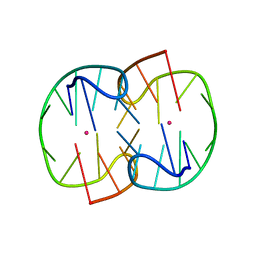 | | The Pb2+ complexed structure of TBA G8C mutant | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*CP*TP*GP*GP*TP*TP*GP*G)-3'), LEAD (II) ION | | Authors: | Liu, H.H, Gao, Y.Q, Sheng, J, Gan, J.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Structure-guided development of Pb 2+ -binding DNA aptamers.
Sci Rep, 12, 2022
|
|
