1MGT
 
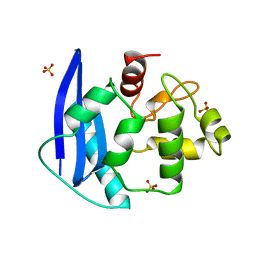 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | Descriptor: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | Authors: | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
5D6U
 
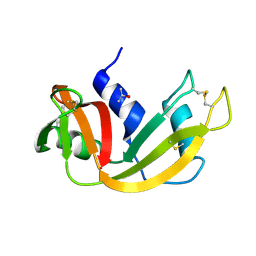 | |
5D97
 
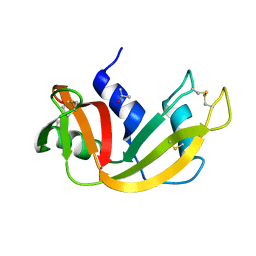 | | Neutron crystal structure of H2O-solvent ribonuclease A | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Chatake, T, Fujiwara, S. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | A technique for determining the deuterium/hydrogen contrast map in neutron macromolecular crystallography
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2N37
 
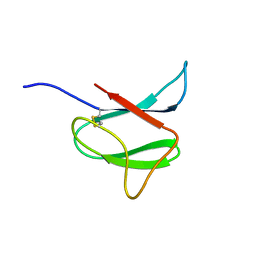 | | Solution structure of AVR-Pia | | Descriptor: | AVR-Pia protein | | Authors: | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
4ZZ4
 
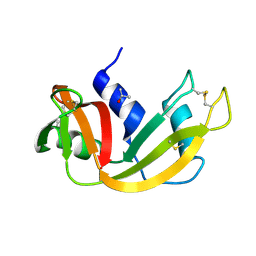 | |
9JF5
 
 | | Arginine decarboxylase in Aspergillus oryzae complexed with arginine | | Descriptor: | 1,2-ETHANEDIOL, AGMATINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9JFN
 
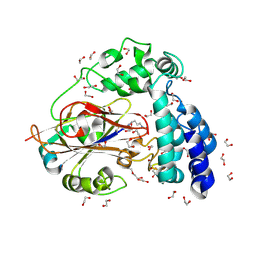 | | Arginine decarboxylase in Aspergillus oryzae complexed with agmatine | | Descriptor: | 1,2-ETHANEDIOL, AGMATINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-05 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9JER
 
 | | Arginine decarboxylase in Aspergillus oryzae, ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-tryptophan decarboxylase PsiD-like domain-containing protein | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
8W48
 
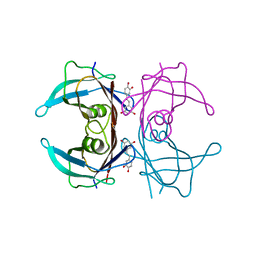 | |
1WNS
 
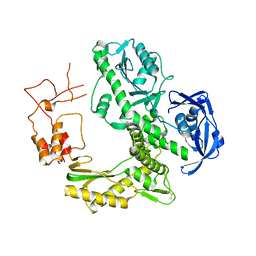 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
7VEI
 
 | | Neutron structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-09-08 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2DKH
 
 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, in complex with the substrate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 3-hydroxybenzoate hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | Deposit date: | 2006-04-11 | | Release date: | 2006-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
2CW7
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2DKI
 
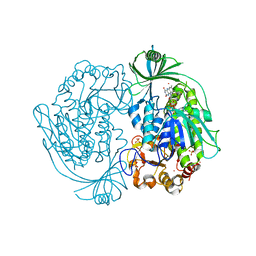 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, under pressure of xenon gas (12 atm) | | Descriptor: | 3-HYDROXYBENZOATE HYDROXYLASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | Deposit date: | 2006-04-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
7FCW
 
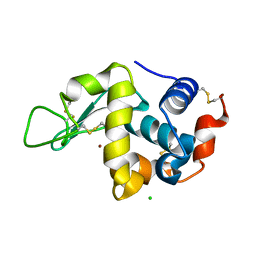 | | X-ray structure of H2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7FCU
 
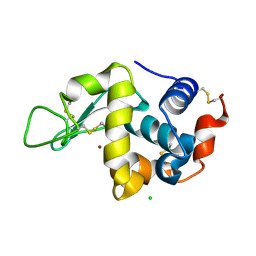 | | X-ray structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
