2FJB
 
 | | Adenosine-5'-phosphosulfate reductase im complex with products | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, ADENOSINE MONOPHOSPHATE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
2FJE
 
 | | adenosine-5-phosphosulfate reductase oxidized state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, adenylylsulfate reductase, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
2FJD
 
 | | adenosine-5-phosphosulfate reductase in complex with sulfite (covalent adduct) | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, IRON/SULFUR CLUSTER, adenylylsulfate reductase, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
2FJA
 
 | | adenosine 5'-phosphosulfate reductase in complex with substrate | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schiffer, A, Fritz, G, Kroneck, P.M, Ermler, U. | | Deposit date: | 2006-01-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of the iron-sulfur flavoenzyme adenosine-5'-phosphosulfate reductase based on the structural characterization of different enzymatic states
Biochemistry, 45, 2006
|
|
8A1V
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1W
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q1 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1T
 
 | | Sodium pumping NADH-quinone oxidoreductase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1X
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1U
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2RGI
 
 | |
3CZT
 
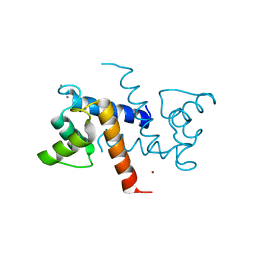 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 9 | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-04-30 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
3D0Y
 
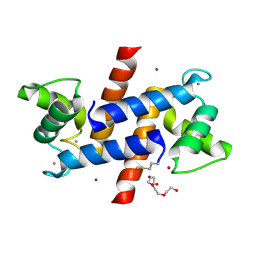 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 6.5 | | Descriptor: | CALCIUM ION, Protein S100-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
3D10
 
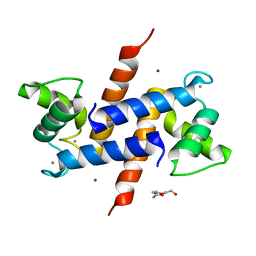 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 10.0 | | Descriptor: | CALCIUM ION, Protein S100-B, TRIETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
3CJJ
 
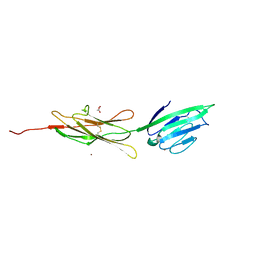 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
2H61
 
 | | X-ray structure of human Ca2+-loaded S100B | | Descriptor: | CALCIUM ION, Protein S100-B, TETRAETHYLENE GLYCOL | | Authors: | Ostendorp, T, Heizmann, C.W, Kroneck, P.M.H, Fritz, G. | | Deposit date: | 2006-05-30 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insights into RAGE activation by multimeric S100B.
Embo J., 26, 2007
|
|
1DUW
 
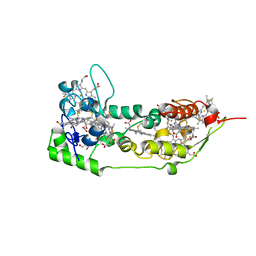 | | STRUCTURE OF NONAHEME CYTOCHROME C | | Descriptor: | GLYCEROL, HEME C, NONAHEME CYTOCHROME C | | Authors: | Umhau, S, Fritz, G, Diederichs, K, Breed, J, Kroneck, P.M, Welte, W. | | Deposit date: | 2000-01-19 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Three-dimensional structure of the nonaheme cytochrome c from Desulfovibrio desulfuricans Essex in the Fe(III) state at 1.89 A resolution.
Biochemistry, 40, 2001
|
|
1KSO
 
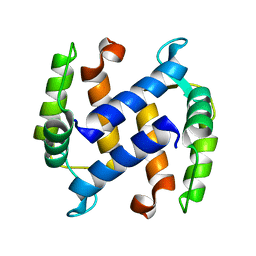 | | CRYSTAL STRUCTURE OF APO S100A3 | | Descriptor: | S100 CALCIUM-BINDING PROTEIN A3 | | Authors: | Mittl, P.R, Fritz, G, Sargent, D.F, Richmond, T.J, Heizmann, C.W, Grutter, M.G. | | Deposit date: | 2002-01-14 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-free MIRAS phasing: structure of apo-S100A3.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1I77
 
 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ESSEX 6 | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Einsle, O, Foerster, S, Mann, K.H, Fritz, G, Messerschmidt, A, Kroneck, P.M.H. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Spectroscopic investigation and determination of reactivity and structure of the tetraheme cytochrome c3 from Desulfovibrio desulfuricans Essex 6.
Eur.J.Biochem., 268, 2001
|
|
1PJX
 
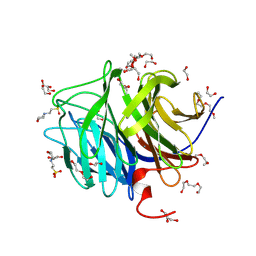 | | 0.85 ANGSTROM STRUCTURE OF SQUID GANGLION DFPASE | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Koepke, J, Rueterjans, H, Luecke, C, Fritzsch, G. | | Deposit date: | 2003-06-04 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Statistical analysis of crystallographic data obtained from squid ganglion DFPase at 0.85 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2BGH
 
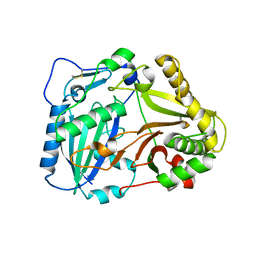 | | Crystal structure of Vinorine Synthase | | Descriptor: | VINORINE SYNTHASE | | Authors: | Ma, X, Koepke, J, Panjikar, S, Fritzsch, G, Stoeckigt, J. | | Deposit date: | 2004-12-22 | | Release date: | 2005-01-24 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Vinorine Synthase, the First Representative of the Bahd Superfamily.
J.Biol.Chem., 280, 2005
|
|
2IAS
 
 | | Crystal structure of squid ganglion DFPase W244F mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2IAX
 
 | | Crystal structure of squid ganglion DFPase D232S mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Katsemi, V, Luecke, C, Koepke, J, Loehr, F, Maurer, S, Fritzsch, G, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme
Biochemistry, 44, 2005
|
|
2IAR
 
 | | Crystal structure of squid ganglion DFPase W244H mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
2IAQ
 
 | | Crystal structure of squid ganglion DFPase S271A mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
