5AFE
 
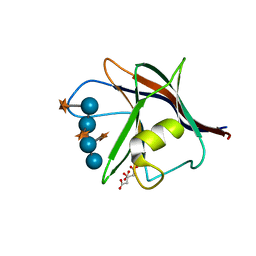 | |
5A8C
 
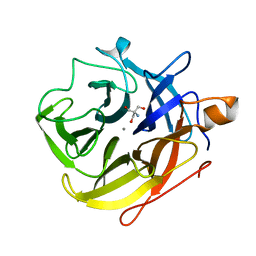 | | The ultra high resolution structure of a novel alpha-L-arabinofuranosidase (CtGH43) from Clostridium thermocellum ATCC 27405 with bound trimethyl N-Oxide (TRS) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Goyal, A, Ahmed, S, Sharma, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-07-14 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Molecular determinants of substrate specificity revealed by the structure of Clostridium thermocellum arabinofuranosidase 43A from glycosyl hydrolase family 43 subfamily 16.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5M2O
 
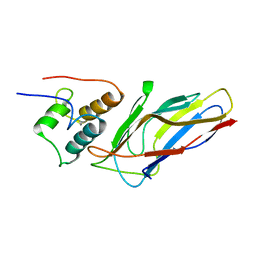 | | R. flavefaciens' third ScaB cohesin in complex with a group 1 dockerin | | Descriptor: | CALCIUM ION, Group I Dockerin, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Najmudin, S, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes.
Sci Rep, 7, 2017
|
|
7QUZ
 
 | | Crystal structure of the SeMet octameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, GLYCEROL | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-01-19 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Bacillus circulans IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
7R1N
 
 | | Crystal structure of the Tetrameric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | Beta-1,3-glucanase bglH, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
7R3T
 
 | | Crystal structure of the Dimeric C-terminal Big_2-CBM56 domains from Paenibacillus illinoisensis (Bacillus circulans IAM1165) beta-1,3-glucanase H | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-1,3-glucanase bglH, CHLORIDE ION, ... | | Authors: | Najmudin, S, Venditto, I, Fontes, C.M.G.A, Bule, P. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural and biochemical characterization of C-terminal Big_2-CBM56 domains of Paenibacillus illinoisensis IAM1165 beta-1,3-glucanase H and Paenibacillus sp CBM56
To be published
|
|
4BA6
 
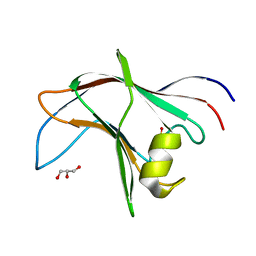 | | High Resolution structure of the C-terminal family 65 Carbohydrate Binding Module (CBM65B) of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | Endoglucanase cel5A, GLYCEROL | | Authors: | Venditto, I, Luis, A.S, Basle, A, Temple, M, Ferreira, L.M.A, Fontes, C.M.G.A, Gilbert, H.J, Najmudin, S. | | Deposit date: | 2012-09-11 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Understanding how noncatalytic carbohydrate binding modules can display specificity for xyloglucan.
J. Biol. Chem., 288, 2013
|
|
2M1J
 
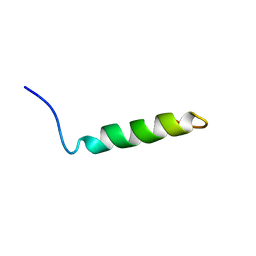 | | Ovine Doppel Signal peptide (1-30) | | Descriptor: | Prion-like protein doppel | | Authors: | Pimenta, J, Viegas, A, Sardinha, J, Santos, A, Cantante, C, Dias, F.M.V, Soares, R, Cabrita, E.J, Fontes, C.M.G.A, Prates, J.A.M, Pereira, R.M.L.N. | | Deposit date: | 2012-11-28 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and SRP54M predicted interaction of the N-terminal sequence (1-30) of the ovine Doppel protein.
Peptides, 49C, 2013
|
|
1WB6
 
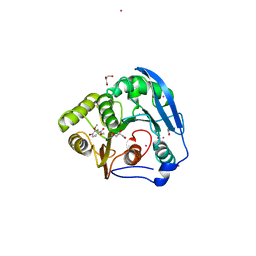 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with vanillate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5NRK
 
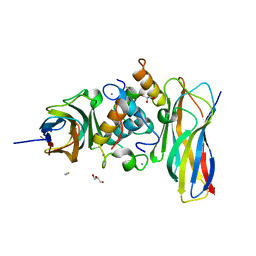 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S15I, I16N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S15I, I16N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
1WB5
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with syringate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5N5P
 
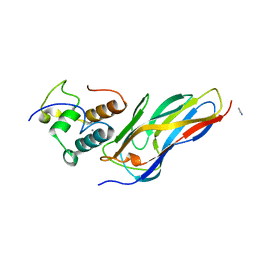 | | Crystal structure of Ruminococcus flavefaciens' type III complex containing the fifth cohesin from scaffoldin B and the dockerin from scaffoldin A | | Descriptor: | ACETONITRILE, CALCIUM ION, Putative cellulosomal scaffoldin protein | | Authors: | Bule, P, Carvalho, A.L, Najmudin, S, Fontes, C.M.G.A. | | Deposit date: | 2017-02-14 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Higher order scaffoldin assembly in Ruminococcus flavefaciens cellulosome is coordinated by a discrete cohesin-dockerin interaction.
Sci Rep, 8, 2018
|
|
4D3L
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 in the orthorhombic form | | Descriptor: | (3S)-3-HYDROXYHEPTANEDIOIC ACID, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-10-22 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4DH2
 
 | | Crystal structure of Coh-OlpC(Cthe_0452)-Doc435(Cthe_0435) complex: A novel type I Cohesin-Dockerin complex from Clostridium thermocellum ATTC 27405 | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin type 1, ... | | Authors: | Alves, V.D, Carvalho, A.L, Najmudin, S.H, Bras, J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Clostridium thermocellum Type I Cohesin-Dockerin Complexes Reveal a Single Binding Mode.
J.Biol.Chem., 287, 2012
|
|
5NRM
 
 | | Crystal structure of the sixth cohesin from Acetivibrio cellulolyticus' scaffoldin B in complex with Cel5 dockerin S51I, L52N mutant | | Descriptor: | CALCIUM ION, DocCel5: Type I dockerin repeat domain from A. cellulolyticus family 5 endoglucanase WP_010249057 S51I, L52N mutant, ... | | Authors: | Bule, P, Najmudin, S, Fontes, C.M.G.A, Alves, V.D. | | Deposit date: | 2017-04-24 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses generate novel specificities to assemble the components of multienzyme bacterial cellulosome complexes.
J. Biol. Chem., 293, 2018
|
|
5A6L
 
 | | High resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with two xylobiose units bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5A6M
 
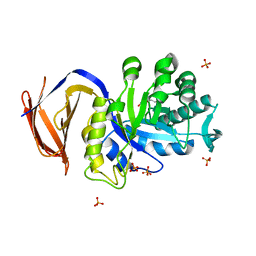 | | Determining the specificities of the catalytic site from the very high resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with a xylotetraose bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5AOZ
 
 | | High resolution SeMet structure of the third cohesin from Ruminococcus flavefaciens scaffoldin protein, ScaB | | Descriptor: | GLYCEROL, PUTATIVE CELLULOSOMAL SCAFFOLDIN PROTEIN | | Authors: | Bule, P, Carvalho, A.L, Santos, H, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural Characterization of the Third Cohesin from Ruminococcus Flavefaciens Scaffoldin Protein, Scab
To be Published
|
|
4AFD
 
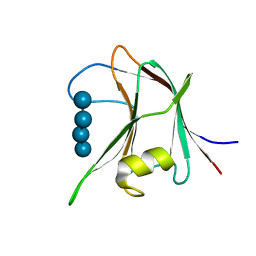 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens with a partially bound cellotetraose moeity. | | Descriptor: | ENDOGLUCANASE CEL5A, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Understanding How Noncatalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
4AFM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens. | | Descriptor: | ACETATE ION, ENDOGLUCANASE CEL5A, GLYCEROL | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
2C4X
 
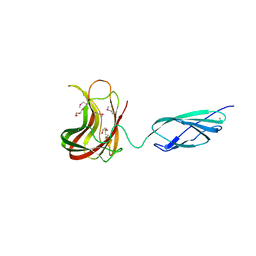 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
4AEK
 
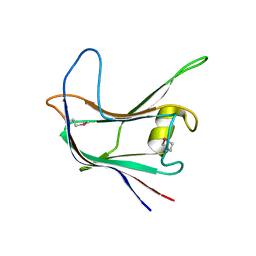 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
2C26
 
 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
4AEM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
4UQA
 
 | | X-ray structure of glucuronoxylan-xylanohydrolase (Xyn30A) from Clostridium thermocellum | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, HISTIDINE | | Authors: | Freire, F, Verma, A.K, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-06-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
