6RCC
 
 | | Domain C P140 Mycoplasma genitalium | | Descriptor: | Adhesin P1, CHLORIDE ION, SODIUM ION | | Authors: | Vizarraga, D, Aparicio, D, Perez, R, Illanes, R, Fita, I. | | Deposit date: | 2019-04-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Alternative conformation of the C-domain of the P140 protein from Mycoplasma genitalium.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6R43
 
 | | Structure of P110 from Mycoplasma Genitalium complexed with 6'-SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Aparicio, D, Fita, I. | | Deposit date: | 2019-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Mycoplasma genitalium adhesin P110 binds sialic-acid human receptors.
Nat Commun, 9, 2018
|
|
3ZZI
 
 | | Crystal structure of a tetrameric acetylglutamate kinase from Saccharomyces cerevisiae | | Descriptor: | ACETYLGLUTAMATE KINASE | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
3ZZG
 
 | | Crystal structure of the amino acid kinase domain from Saccharomyces cerevisiae acetylglutamate kinase without ligands | | Descriptor: | ACETYLGLUTAMATE KINASE | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
3P9P
 
 | | Structure of I274V variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3P9Q
 
 | | Structure of I274C variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3P9S
 
 | | Structure of I274A variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3P9R
 
 | | Structure of I274G variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ8
 
 | | Structure of I274C variant of E. coli KatE[] - Images 37-42 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ4
 
 | | Structure of I274C variant of E. coli KatE[] - Images 13-18 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ2
 
 | | Structure of I274C variant of E. coli KatE[] - Images 1-6 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
1QWM
 
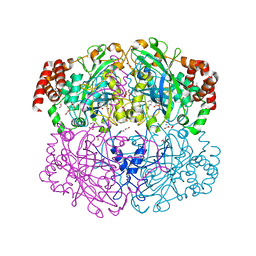 | | Structure of Helicobacter pylori catalase with formic acid bound | | Descriptor: | AZIDE ION, FORMIC ACID, KatA catalase, ... | | Authors: | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Odenbreit, S, Nicholls, P, Fita, I. | | Deposit date: | 2003-09-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
3PQ3
 
 | | Structure of I274C variant of E. coli KatE[] - Images 7-12 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ7
 
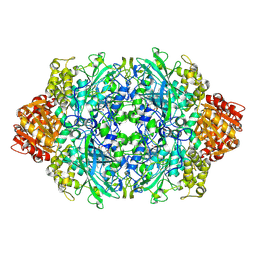 | | Structure of I274C variant of E. coli KatE[] - Images 31-36 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
1GMI
 
 | | Structure of the c2 domain from novel protein kinase C epsilon | | Descriptor: | MAGNESIUM ION, PROTEIN KINASE C, EPSILON TYPE | | Authors: | Ochoa, W.F, Garcia-Garcia, J, Fita, I, Corbalan-Garcia, S, Verdaguer, N, Gomez-Fernandez, J.C. | | Deposit date: | 2001-09-14 | | Release date: | 2001-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the C2 Domain from Novel Protein Kinase Cepsilon. A Membrane Binding Model for Ca(2+ )-Independent C2 Domains
J.Mol.Biol., 311, 2001
|
|
3PQ6
 
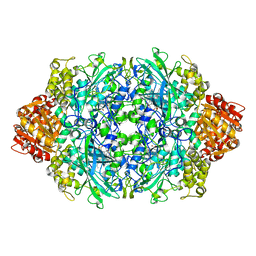 | | Structure of I274C variant of E. coli KatE[] - Images 25-30 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3PQ5
 
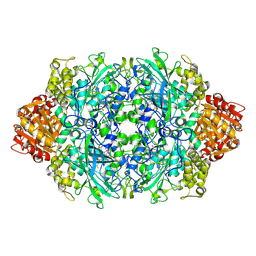 | | Structure of I274C variant of E. coli KatE[] - Images 19-24 | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
1GS5
 
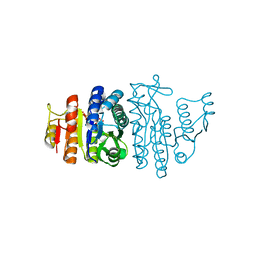 | | N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate N-acetylglutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2001-12-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
1GSJ
 
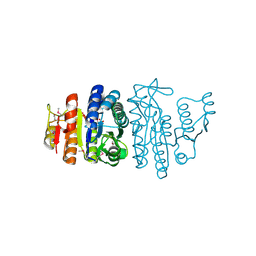 | | Selenomethionine substituted N-acetyl-L-glutamate kinase from Escherichia coli complexed with its substrate n-acetyl-L-glutamate and its substrate analog AMPPNP | | Descriptor: | ACETYLGLUTAMATE KINASE, MAGNESIUM ION, N-ACETYL-L-GLUTAMATE, ... | | Authors: | Ramon-Maiques, S, Marina, A, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Acetylglutamate Kinase, a Key Enzyme for Arginine Biosynthesis and a Prototype for the Amino Acid Kinase Enzyme Family, During Catalysis
Structure, 10, 2002
|
|
6RNN
 
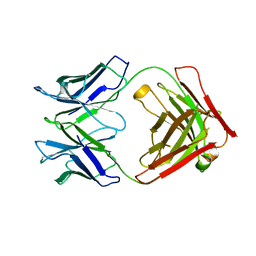 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin light chain | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6RQG
 
 | |
2BFW
 
 | | Structure of the C domain of glycogen synthase from Pyrococcus abyssi | | Descriptor: | ACETATE ION, GLGA GLYCOGEN SYNTHASE, SULFATE ION | | Authors: | Horcajada, C, Guinovart, J.J, Fita, I, Ferrer, J.C. | | Deposit date: | 2004-12-15 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Archaeal Glycogen Synthase: Insights Into Oligomerisation and Substrate Binding of Eukaryotic Glycogen Synthases.
J.Biol.Chem., 281, 2006
|
|
1QWS
 
 | | Structure of the D181N variant of catalase HPII from E. coli | | Descriptor: | Catalase HPII, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chelikani, P, Carpena, X, Fita, I, Loewen, P.C. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An electrical potential in the access channel of catalases enhances catalysis
J.Biol.Chem., 278, 2003
|
|
2D47
 
 | | MOLECULAR STRUCTURE OF A COMPLETE TURN OF A-DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*GP*CP*GP*GP*GP*GP*G)-3'), SPERMINE | | Authors: | Verdaguer, N, Aymami, J, Fernandez-Forner, D, Fita, I, Coll, M, Huynh-Dinh, T, Igolen, J, Subirana, J.A. | | Deposit date: | 1991-10-02 | | Release date: | 1991-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of a complete turn of A-DNA.
J.Mol.Biol., 221, 1991
|
|
4F0X
 
 | | Crystal structure of human Malonyl-CoA Decarboxylase (Peroxisomal Isoform) | | Descriptor: | Malonyl-CoA decarboxylase, mitochondrial, N~3~-[(2R)-2-hydroxy-4-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-3,3-dimethylbutanoyl]-beta-alaninamide | | Authors: | Aparicio, D, Perez, R, Fita, I. | | Deposit date: | 2012-05-05 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Asymmetry and Disulfide Bridges among Subunits Modulate the Activity of Human Malonyl-CoA Decarboxylase.
J.Biol.Chem., 288, 2013
|
|
