4FHH
 
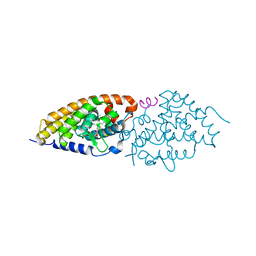 | | Development of synthetically accessible non-secosteroidal hybrid molecules combining vitamin D receptor agonism and histone deacetylase inhibition | | Descriptor: | N-hydroxy-2-{4-[3-(4-{[(2S)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetamide, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Fischer, J, Wang, T.T, Kaldre, D, Rochel, N, Moras, D, White, J.H, Gleason, J.L. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Synthetically accessible non-secosteroidal hybrid molecules combining vitamin d receptor agonism and histone deacetylase inhibition.
Chem.Biol., 19, 2012
|
|
4FHI
 
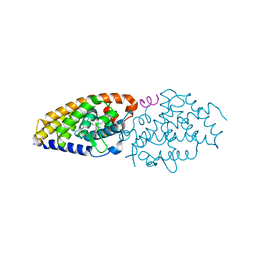 | | Development of synthetically accessible non-secosteroidal hybrid molecules combining vitamin D receptor agonism and histone deacetylase inhibition | | Descriptor: | N-hydroxy-2-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetamide, SRC-1, Vitamin Nuclear Receptor | | Authors: | Fischer, J, Wang, T.T, Kaldre, D, Rochel, N, Moras, D, White, J.H, Gleason, J.L. | | Deposit date: | 2012-06-06 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetically accessible non-secosteroidal hybrid molecules combining vitamin d receptor agonism and histone deacetylase inhibition.
Chem.Biol., 19, 2012
|
|
6QSO
 
 | |
6QSL
 
 | |
6QSM
 
 | |
4Q9W
 
 | |
4Q9X
 
 | | mTFP* PdCl2 soak | | Descriptor: | CARBONATE ION, CHLORIDE ION, GFP-like fluorescent chromoprotein cFP484, ... | | Authors: | Fischer, J, Quitterer, F, Groll, M, Eppinger, J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | mTFP*: An imperishable and versatile protein host for anchoring diverse ligands and organocatalysts
To be Published
|
|
4R6D
 
 | |
6E0I
 
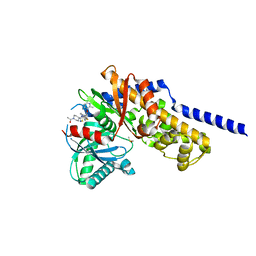 | | Crystal structure of Glucokinase in complex with compound 72 | | Descriptor: | 1-{4-[5-({3-[(2-methylpyridin-3-yl)oxy]-5-[(pyridin-2-yl)sulfanyl]pyridin-2-yl}amino)-1,2,4-thiadiazol-3-yl]piperidin-1 -yl}ethan-1-one, DIMETHYL SULFOXIDE, Glucokinase, ... | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
6E0E
 
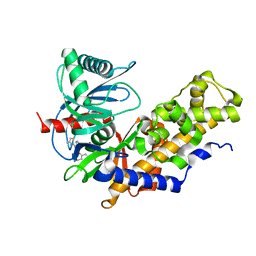 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
1VTQ
 
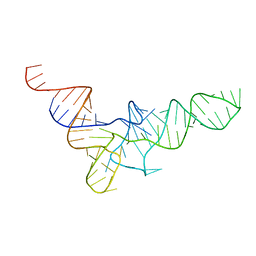 | | THREE-DIMENSIONAL STRUCTURE OF YEAST T-RNA-ASP. I. STRUCTURE DETERMINATION | | Descriptor: | T-RNA-ASP | | Authors: | Comarmond, M.B, Giege, R, Thierry, J.C, Moras, D, Fischer, J. | | Deposit date: | 1985-06-11 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of Yeast T-RNA-ASP. I. Structure Determination
Acta Crystallogr.,Sect.B, 42, 1986
|
|
5CWV
 
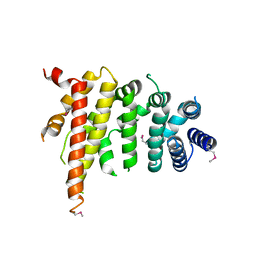 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWU
 
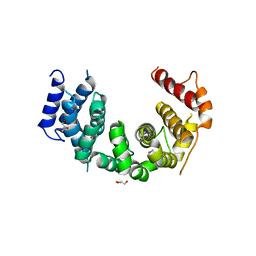 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWW
 
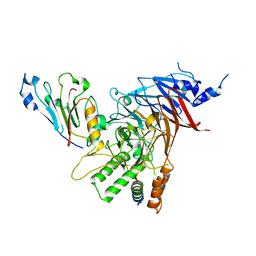 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
