6H3E
 
 | | Receptor-bound Ghrelin conformation | | Descriptor: | Appetite-regulating hormone, octan-1-amine | | Authors: | Ferre, G, Damian, M, M'Kadmi, C, Saurel, O, Czaplicki, G, Demange, P, Marie, J, Fehrentz, J.A, Baneres, J.L, Milon, A. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4A1M
 
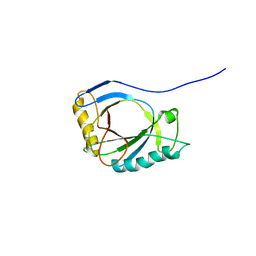 | | NMR Structure of protoporphyrin-IX bound murine p22HBP | | Descriptor: | HEME-BINDING PROTEIN 1 | | Authors: | Goodfellow, B.J, Dias, J.S, Macedo, A.L, Ferreira, G.C, Peterson, F.C, Volkman, B.F, Duarte, I.C.N. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of Protoporphyrin-Ix Bound Murine P22Hbp
To be Published
|
|
1H7D
 
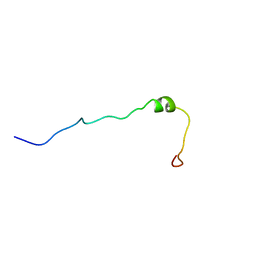 | | Solution structure of the 49 aa presequence of 5-ALAS | | Descriptor: | AMINOLEVULINIC ACID SYNTHASE 2, ERYTHROID | | Authors: | Goodfellow, B.J, Dias, J.S, Ferreira, G.C, Wray, V, Henklein, P, Macedo, A.L. | | Deposit date: | 2001-07-05 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and Heme Binding of the Presequence of Murine 5-Aminolevulinate Synthase
FEBS Lett., 505, 2001
|
|
1H7J
 
 | | Solution structure of the 26 aa presequence of 5-ALAS | | Descriptor: | AMINOLEVULINIC ACID SYNTHASE 2, ERYTHROID | | Authors: | Goodfellow, B.J, Dias, J.S, Ferreira, G.C, Wray, V, Henklein, P, Macedo, A.L. | | Deposit date: | 2001-07-08 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and Heme Binding of the Presequence of Murine 5-Aminolevulinate Synthase
FEBS Lett., 505, 2001
|
|
2AC4
 
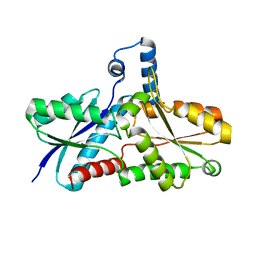 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2AC2
 
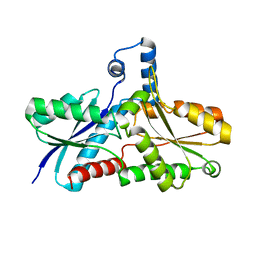 | | Crystal structure of the Tyr13Phe mutant variant of Bacillus subtilis Ferrochelatase with Zn(2+) bound at the active site | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
