3E96
 
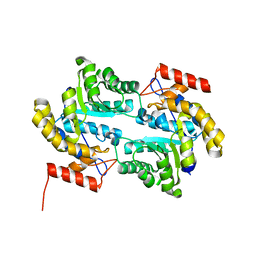 | | Crystal structure of dihydrodipicolinate synthase from bacillus clausii | | Descriptor: | Dihydrodipicolinate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase from bacillus clausii
To be Published
|
|
3NWR
 
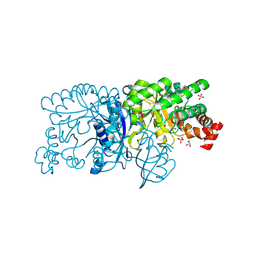 | | Crystal structure of a rubisco-like protein from Burkholderia fungorum | | Descriptor: | A rubisco-like protein, GLYCEROL, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-07-10 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of a rubisco-like protein from Burkholderia fungorum
To be Published
|
|
3NQ7
 
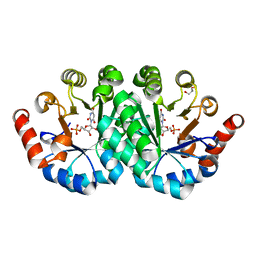 | | Crystal structure of the mutant F71A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-06-29 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Crystal structure of the mutant F71A of orotidine 5'-monophosphate
decarboxylase from Methanobacterium thermoautotrophicum complexed with
inhibitor BMP
To be Published
|
|
3P3B
 
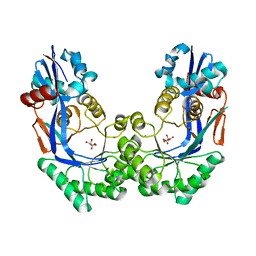 | | CRYSTAL STRUCTURE OF Galacturonate DEHYDRATASE FROM GEOBACILLUS SP. COMPLEXED WITH D-TARTRATE | | Descriptor: | D(-)-TARTARIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | CRYSTAL STRUCTURE OF Galacturonate DEHYDRATASE FROM GEOBACILLUS SP. COMPLEXED WITH D-TARTRATE.
To be Published
|
|
3P0W
 
 | | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate | | Descriptor: | D-GLUCARATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of D-Glucarate dehydratase from Ralstonia solanacearum complexed with Mg and D-glucarate
To be Published
|
|
3M5Z
 
 | | Crystal structure of the mutant V182A,I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3MDU
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | Descriptor: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4003 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3OZY
 
 | | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg and m-Xylarate | | Descriptor: | ACETIC ACID, D-xylaric acid, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2994 Å) | | Cite: | Crystal structure of enolase superfamily member from Bordetella bronchiseptica complexed with Mg and m-Xylarate
To be Published
|
|
3GD6
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate | | Descriptor: | Muconate cycloisomerase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate.
To be Published
|
|
3M1Z
 
 | | Crystal structure of the mutant V182A.V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3M44
 
 | | Crystal structure of the mutant V201A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3PDW
 
 | | Crystal structure of putative p-nitrophenyl phosphatase from Bacillus subtilis | | Descriptor: | ACETIC ACID, GLYCEROL, Uncharacterized hydrolase yutF | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.596 Å) | | Cite: | Crystal structure of putative p-nitrophenyl phosphatase from Bacillus subtilis
To be Published
|
|
3MTW
 
 | | Crystal structure of L-Lysine, L-Arginine carboxypeptidase Cc2672 from Caulobacter Crescentus CB15 complexed with N-methyl phosphonate derivative of L-Arginine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L-Arginine carboxypeptidase Cc2672, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-05-01 | | Release date: | 2010-07-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Identification and Structure Determination of Two Novel Prolidases from cog1228 in the Amidohydrolase Superfamily
Biochemistry, 49, 2010
|
|
3GDM
 
 | |
3M7V
 
 | | Crystal structure of phosphopentomutase from streptococcus mutans | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Fedorov, A.A, Bonanno, J, Fedorov, E.V, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopentomutase from streptococcus mutans
To be Published
|
|
3PWG
 
 | | Crystal structure of the mutant S29G.P34A of D-Glucarate dehydratase from Escherichia coli complexed with product 5-keto-4-deoxy-D-Glucarate | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-12-08 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the mutant S29G.P34A of D-Glucarate dehydratase from Escherichia Coli complexed with product 5-keto-4-deoxy-D-Glucarate
To be Published
|
|
3DTC
 
 | | Crystal structure of mixed-lineage kinase MLK1 complexed with compound 16 | | Descriptor: | 12-(2-hydroxyethyl)-2-(1-methylethoxy)-13,14-dihydronaphtho[2,1-a]pyrrolo[3,4-c]carbazol-5(12H)-one, Mitogen-activated protein kinase kinase kinase 9, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, S.L, Hudkins, R.L, Almo, S.C. | | Deposit date: | 2008-07-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mixed-lineage kinase 1 and mixed-lineage kinase 3 subtype-selective dihydronaphthyl[3,4-a]pyrrolo[3,4-c]carbazole-5-ones: optimization, mixed-lineage kinase 1 crystallography, and oral in vivo activity in 1-methyl-4-phenyltetrahydropyridine models.
J.Med.Chem., 51, 2008
|
|
3PWI
 
 | | Crystal structure of the mutant P34A of D-Glucarate dehydratase from Escherichia coli complexed with product 5-keto-4-deoxy-D-Glucarate | | Descriptor: | 2,3-DIHYDROXY-5-OXO-HEXANEDIOATE, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-12-08 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2296 Å) | | Cite: | Crystal structure of the mutant P34A of D-Glucarate dehydratase from Escherichia Coli complexed with product 5-keto-4-deoxy-D-Glucarate
To be Published
|
|
3NEH
 
 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3PFR
 
 | | Crystal structure of D-Glucarate dehydratase related protein from Actinobacillus Succinogenes complexed with D-Glucarate | | Descriptor: | D-GLUCARATE, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Mills-Groninger, F, Ghasempur, S, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of D-Glucarate dehydratase related protein from Actinobacillus Succinogenes complexed with D-Glucarate
To be Published
|
|
3M3P
 
 | | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus | | Descriptor: | Glutamine amido transferase | | Authors: | Fedorov, A.A, Domagalski, M, Fedorov, E.V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of glutamine amido transferase from Methylobacillus Flagellatus
To be Published
|
|
3M5X
 
 | | Crystal structure of the mutant V182A,I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
2AJT
 
 | | Crystal structure of L-Arabinose Isomerase from E.coli | | Descriptor: | L-arabinose isomerase | | Authors: | Manjasetty, B.A, Fedorov, E.V, Almo, S.C, Chance, M.R, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-02 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli L-Arabinose Isomerase (ECAI), The Putative Target of Biological Tagatose Production
J.Mol.Biol., 360, 2006
|
|
2OOG
 
 | | Crystal structure of glycerophosphoryl diester phosphodiesterase from Staphylococcus aureus | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SULFATE ION, ... | | Authors: | Patskovsky, Y, Fedorov, E, Toro, R, Sauder, J.M, Smith, D, Freeman, J, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Glycerophosphoryl Diester Phosphodiesterase from Staphylococcus Aureus
To be Published
|
|
