7REQ
 
 | |
1REQ
 
 | | METHYLMALONYL-COA MUTASE | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, GLYCEROL, ... | | Authors: | Evans, P.R, Mancia, F. | | Deposit date: | 1996-01-19 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution.
Structure, 4, 1996
|
|
1URU
 
 | | Amphiphysin BAR domain from Drosophila | | Descriptor: | AMPHIPHYSIN | | Authors: | Evans, P.R, Kent, H.M. | | Deposit date: | 2003-11-06 | | Release date: | 2003-12-04 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bar Domains as Sensors of Membrane Curvature: The Amphiphysin Bar Structure
Science, 303, 2004
|
|
1E1C
 
 | | METHYLMALONYL-COA MUTASE H244A Mutant | | Descriptor: | COBALAMIN, DESULFO-COENZYME A, METHYLMALONYL-COA MUTASE ALPHA CHAIN, ... | | Authors: | Evans, P.R, Thoma, N.H. | | Deposit date: | 2000-04-30 | | Release date: | 2000-05-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Protection of Radical Intermediates at the Active Site of Adenosylcobalamin-Dependent Methylmalonyl-Coa Mutase
Biochemistry, 39, 2000
|
|
1GYW
 
 | | Gamma-adaptin appendage domain from clathrin adaptor AP1 A753D mutant | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, CHLORIDE ION | | Authors: | Evans, P.R, Miele, A.E, Owen, D.J, McMahon, H.M, Kent, H.M. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin.
Structure, 10, 2002
|
|
1GYV
 
 | | Gamma-adaptin appendage domain from clathrin adaptor AP1, L762E mutant | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT | | Authors: | Evans, P.R, Owen, D.J, McMahon, H.M, Kent, H.M. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin.
Structure, 10, 2002
|
|
1GYU
 
 | |
6PFK
 
 | |
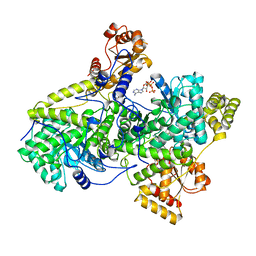
2REQ
 
 | |
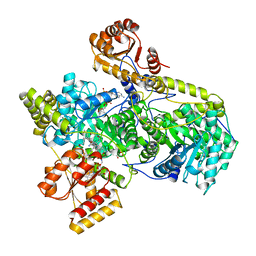
6REQ
 
 | |
2VX8
 
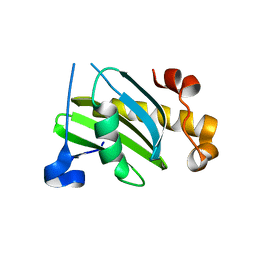 | | Vamp7 longin domain Hrb peptide complex | | Descriptor: | CHLORIDE ION, NUCLEOPORIN-LIKE PROTEIN RIP, VESICLE-ASSOCIATED MEMBRANE PROTEIN 7 | | Authors: | Evans, P.R, Owen, D.J, Luzio, J.P. | | Deposit date: | 2008-07-01 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Sorting of the Snare Vamp7 Into Endocytic Clathrin-Coated Vesicles by the Arfgap Hrb.
Cell(Cambridge,Mass.), 134, 2008
|
|
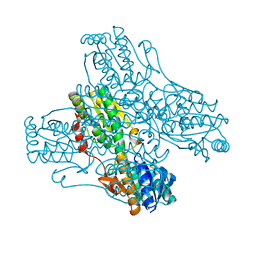
3PFK
 
 | |
4PFK
 
 | | PHOSPHOFRUCTOKINASE. STRUCTURE AND CONTROL | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Evans, P.R, Hudson, P.J. | | Deposit date: | 1988-01-25 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphofructokinase: structure and control.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
3REQ
 
 | |
4REQ
 
 | | Methylmalonyl-COA Mutase substrate complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GLYCEROL, ... | | Authors: | Evans, P.R, Mancia, F. | | Deposit date: | 1998-06-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism.
Structure, 6, 1998
|
|
5REQ
 
 | | Methylmalonyl-COA MUTASE, Y89F Mutant, substrate complex | | Descriptor: | COBALAMIN, GLYCEROL, METHYLMALONYL(CARBADETHIA)-COENZYME A, ... | | Authors: | Evans, P.R, Thomae, N.H. | | Deposit date: | 1998-08-03 | | Release date: | 1998-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of radical intermediates by an active-site tyrosine residue in methylmalonyl-CoA mutase.
Biochemistry, 37, 1998
|
|
1HES
 
 | |
1HFA
 
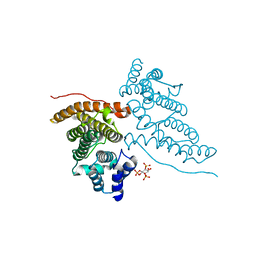 | | CALM-N N-terminal domain of clathrin assembly lymphoid myeloid leukaemia protein, PI(4,5)P2 complex | | Descriptor: | CLATHRIN ASSEMBLY PROTEIN SHORT FORM, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Ford, M.G.J, Evans, P.R, McMahon, H.T. | | Deposit date: | 2000-11-30 | | Release date: | 2001-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Simultaneous Binding of Ptdins(4,5)P2 and Clathrin by Ap180 in the Nucleation of Clathrin Lattices on Membranes
Science, 291, 2001
|
|
1HF8
 
 | |
2PFK
 
 | |
1H0A
 
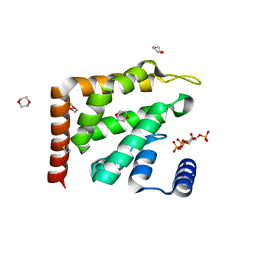 | | Epsin ENTH bound to Ins(1,4,5)P3 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, EPSIN | | Authors: | Ford, M.G.J, McMahon, H.T, Evans, P.R. | | Deposit date: | 2002-06-12 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Curvature of Clathrin-Coated Pits Driven by Epsin
Nature, 419, 2002
|
|
5FJZ
 
 | |
5FK0
 
 | |
5FJW
 
 | |
5FJX
 
 | |
