6SMU
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,12) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SKN
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | Gag protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-26 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SKK
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | Descriptor: | capsid protein | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-26 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5N5S
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase 21 (ALDH21), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
7QG7
 
 | | SARS-CoV-2 macrodomain Nsp3b bound to the remdesivir nucleoside GS-441524 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolane-2-carbonitrile, 1,2-ETHANEDIOL, Papain-like protease nsp3 | | Authors: | Wollenhaupt, J, Linhard, V, Sreeramulu, S, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
7QZO
 
 | | Crystal structure of GacS D1 domain | | Descriptor: | CADMIUM ION, GLYCEROL, Histidine kinase | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
7QZ2
 
 | | Crystal structure of GacS D1 domain in complex with BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CADMIUM ION, Histidine kinase, ... | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
4YMH
 
 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
4YMG
 
 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | Authors: | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
8R0S
 
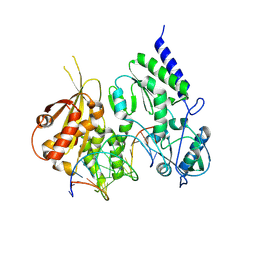 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 2024
|
|
4Z8E
 
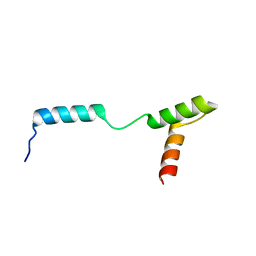 | | TEAD DBD mutant -deltaL1 | | Descriptor: | Transcriptional enhancer factor TEF-1 | | Authors: | Lee, D.-S, Albarado, D.C, Vonrhein, C, Raman, C.S, Veeraraghavan, S. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | A Potential Structural Switch for Regulating DNA-Binding by TEAD Transcription Factors.
J.Mol.Biol., 428, 2016
|
|
3OJO
 
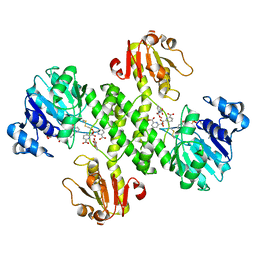 | | Derivative structure of the UDP-N-acetyl-mannosamine dehydrogenase Cap5O from S. aureus | | Descriptor: | Cap5O, EUROPIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Nessler, S, Gruszczyk, J, Olivares-Illana, V, Meyer, P, Morera, S. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure Analysis of the Staphylococcus aureus UDP-N-acetyl-mannosamine Dehydrogenase Cap5O Involved in Capsular Polysaccharide Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
3PAW
 
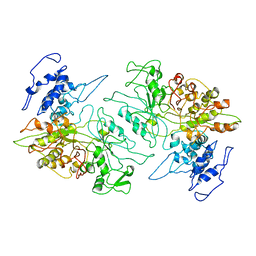 | |
4ZEB
 
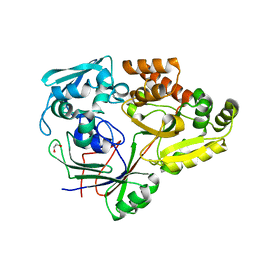 | | PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4ZEK
 
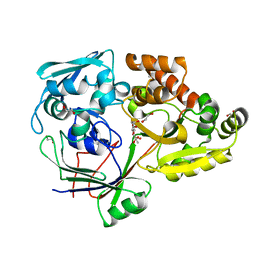 | |
4ZE9
 
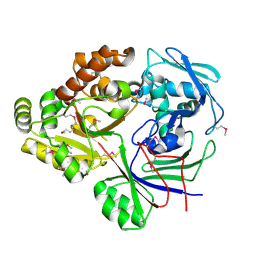 | | Se-PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 2-O-phosphono-alpha-L-arabinopyranose, ABC transporter substrate-binding protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | El Sahili, A, Guimaraes, B.G, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
4ZED
 
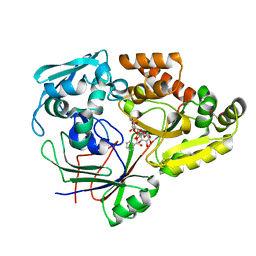 | |
3OJL
 
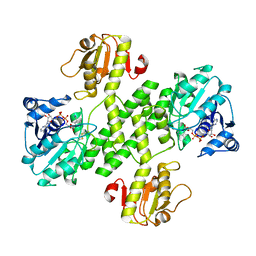 | | Native structure of the UDP-N-acetyl-mannosamine dehydrogenase Cap5O from Staphylococcus aureus | | Descriptor: | Cap5O, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Nessler, S, Gruszczyk, J, Olivares-Illana, V, Meyer, P, Morera, S, Grangeasse, C, Fleurie, A. | | Deposit date: | 2010-08-23 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure Analysis of the Staphylococcus aureus UDP-N-acetyl-mannosamine Dehydrogenase Cap5O Involved in Capsular Polysaccharide Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
4ZA6
 
 | |
7SR2
 
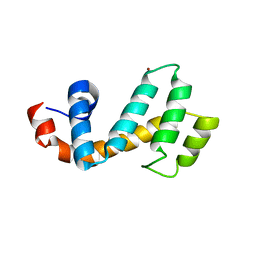 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
7SR1
 
 | |
8A8J
 
 | | Complex of RecF and DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A93
 
 | | Complex of RecF-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6JMI
 
 | | Crystal structure of M.tuberculosis Rv0081 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv0081 | | Authors: | Kumar, A, Phulera, S, Mande, C.S. | | Deposit date: | 2019-03-11 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Structural basis of hypoxic gene regulation by the Rv0081 transcription factor of Mycobacterium tuberculosis.
Febs Lett., 593, 2019
|
|
1G6Y
 
 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM YEAST SACCHAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
