3A8Q
 
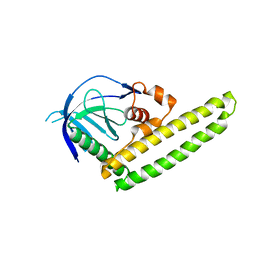 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 2 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 2 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8N
 
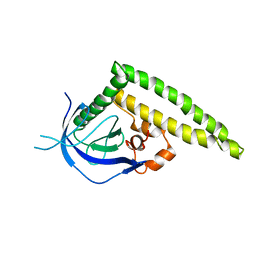 | | Crystal structure of the Tiam1 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 1 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
7FCR
 
 | |
7FCS
 
 | |
1QKJ
 
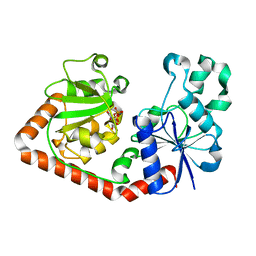 | | T4 Phage B-Glucosyltransferase, Substrate Binding and Proposed Catalytic Mechanism | | 分子名称: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | 著者 | Morera, S, Imberty, I, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | 登録日 | 1999-07-22 | | 公開日 | 1999-07-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | T4 Phage Beta-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism
J.Mol.Biol., 292, 1999
|
|
4K8M
 
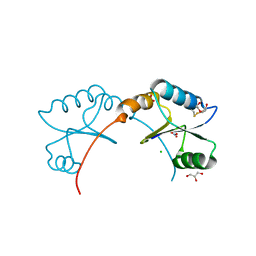 | | High resolution structure of M.tb NRDH | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Phulera, S, Mande, S.C. | | 登録日 | 2013-04-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (0.87 Å) | | 主引用文献 | The crystal structure of Mycobacterium tuberculosis NrdH at 0.87 angstrom suggests a possible mode of its activity.
Biochemistry, 52, 2013
|
|
4K1G
 
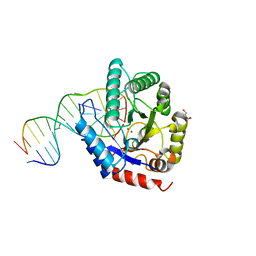 | |
1NUE
 
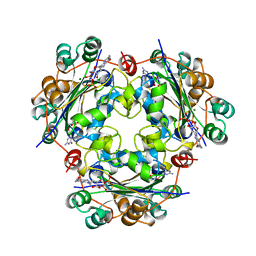 | | X-RAY STRUCTURE OF NM23 HUMAN NUCLEOSIDE DIPHOSPHATE KINASE B COMPLEXED WITH GDP AT 2 ANGSTROMS RESOLUTION | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | 著者 | Morera, S, Lacombe, M.-L, Yingwu, X, Lebras, G, Janin, J. | | 登録日 | 1995-10-06 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 A resolution.
Structure, 3, 1995
|
|
4EUO
 
 | | Structure of Atu4243-GABA sensor | | 分子名称: | ABC transporter, substrate binding protein (Polyamine), GAMMA-AMINO-BUTANOIC ACID, ... | | 著者 | Morera, S, Planamente, S. | | 登録日 | 2012-04-25 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
4EQ7
 
 | | Structure of Atu4243-GABA receptor | | 分子名称: | ABC transporter, substrate binding protein (Polyamine), GLYCEROL, ... | | 著者 | Morera, S, Planamente, S. | | 登録日 | 2012-04-18 | | 公開日 | 2012-11-21 | | 最終更新日 | 2012-12-19 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
1DBD
 
 | |
3FKB
 
 | | Structure of NDPK H122G and tenofovir-diphosphate | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Morera, S, Chen, Y.X. | | 登録日 | 2008-12-16 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Nucleoside diphosphate kinase and the activation of antiviral phosphonate analogs of nucleotides: binding mode and phosphorylation of tenofovir derivatives
Nucleosides Nucleotides Nucleic Acids, 28, 2009
|
|
3RX5
 
 | | structure of AaCel9A in complex with cellotriose-like isofagomine | | 分子名称: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl 4-O-beta-D-glucopyranosyl-beta-D-glucopyranoside, CALCIUM ION, Cellulase, ... | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2011-05-10 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | A fortuitous binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidase
Org.Biomol.Chem., 9, 2011
|
|
3RX7
 
 | | Structure of AaCel9A in complex with cellotetraose-like isofagomine | | 分子名称: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, CALCIUM ION, ... | | 著者 | Morera, S. | | 登録日 | 2011-05-10 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
4MLA
 
 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | 分子名称: | 1,2-ETHANEDIOL, Cytokinin oxidase 2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | 登録日 | 2013-09-06 | | 公開日 | 2015-03-11 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
1FAC
 
 | |
3FJO
 
 | | Structure of chimeric YH CPR | | 分子名称: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | 著者 | Morera, S, Aigrain, L, Truan, G. | | 登録日 | 2008-12-15 | | 公開日 | 2009-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase
Embo Rep., 10, 2009
|
|
4ML8
 
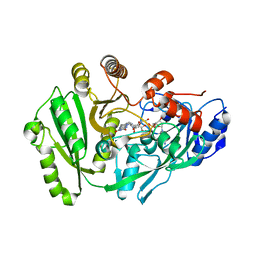 | | Structure of maize cytokinin oxidase/dehydrogenase 2 (ZmCKO2) | | 分子名称: | Cytokinin oxidase 2, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Morera, S, Kopecny, D, Briozzo, P, Koncitikova, R. | | 登録日 | 2013-09-06 | | 公開日 | 2015-03-11 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3RX8
 
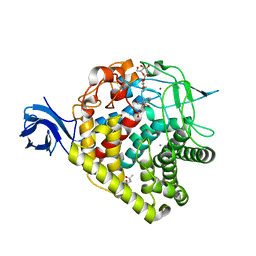 | | structure of AaCel9A in complex with cellobiose-like isofagomine | | 分子名称: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | 著者 | Morera, S. | | 登録日 | 2011-05-10 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Fortuitious binding of inhibitors-derived isofagomine for inverting GH9 beta-glycosidases
Org.Biomol.Chem., 9, 2011
|
|
4E9E
 
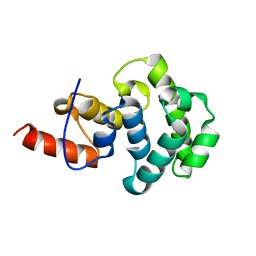 | | Structure of the glycosylase domain of MBD4 | | 分子名称: | Methyl-CpG-binding domain protein 4 | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2012-03-21 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
4E9H
 
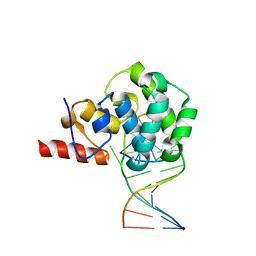 | | structure of glycosylase domain of MBD4 bound to 5hmU containing DNA | | 分子名称: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(5HU)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2012-03-21 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
4EVH
 
 | |
4EA5
 
 | | Structure of the glycoslyase domain of MBD4 bound to a 5hmU containing DNA | | 分子名称: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(5HU)*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2012-03-22 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
4E9G
 
 | | structure of the glycosylase domain of MBD4 bound to thymine containing DNA | | 分子名称: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | 著者 | Morera, S, Vigouroux, A. | | 登録日 | 2012-03-21 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA.
Nucleic Acids Res., 40, 2012
|
|
