7T8T
 
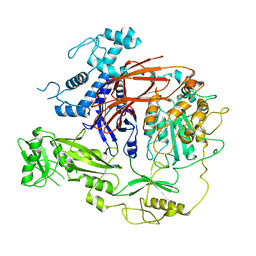 | | CryoEM structure of PLCg1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma, CALCIUM ION | | Authors: | Endo-Streeter, S, Sondek, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | CryoEM structure of PLCg1
To Be Published
|
|
7SQ2
 
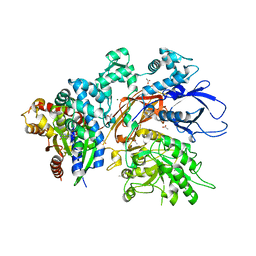 | | Reprocessed and refined structure of Phospholipase C-beta and Gq signaling complex | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, ACETATE ION, CALCIUM ION, ... | | Authors: | Endo-Streeter, S.T, Sondek, J, Harden, T.K. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic Scaffolding Mediated by a Phospholipase C-{beta} and Gq Signaling Complex
Science, 330, 2010
|
|
4FRF
 
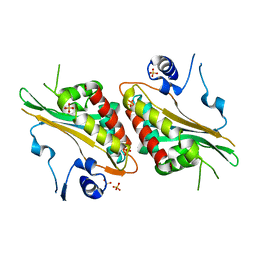 | | Structural Studies and Protein Engineering of Inositol Phosphate Multikinase | | Descriptor: | Inositol polyphosphate multikinase alpha, SULFATE ION | | Authors: | Endo-Streeter, S.T, Tsui, M, Odom, A.R, York, J.D. | | Deposit date: | 2012-06-26 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies and protein engineering of inositol phosphate multikinase.
J.Biol.Chem., 287, 2012
|
|
6WRR
 
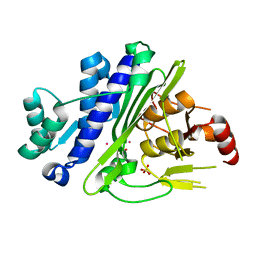 | |
6WRY
 
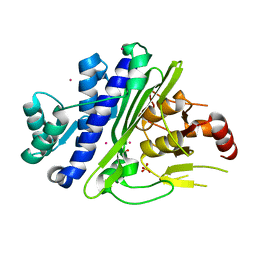 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.R, Endo-Streeter, S, York, J.D. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
6X25
 
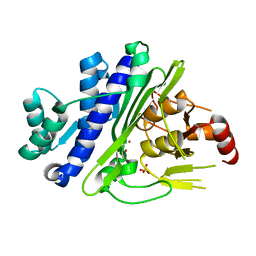 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE INPP1 IN COMPLEX GADOLINIUM AFTER ADDITION OF INOSITOL 1,3,4-TRISPHOSPHATE AND LITHIUM AT 3.2 ANGSTROM RESOLUTION | | Descriptor: | GADOLINIUM ATOM, Inositol polyphosphate 1-phosphatase, SULFATE ION | | Authors: | Dollins, D.E, Endo-Streeter, S, Ren, Y, York, J.D. | | Deposit date: | 2020-05-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for lithium and substrate binding of an inositide phosphatase.
J.Biol.Chem., 296, 2020
|
|
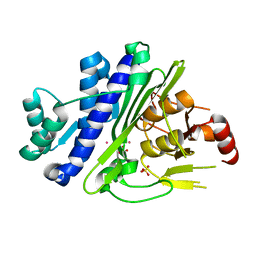
6WRO
 
 | |
4HNW
 
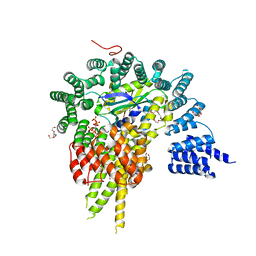 | | The NatA Acetyltransferase Complex Bound To Inositol Hexakisphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HNY
 
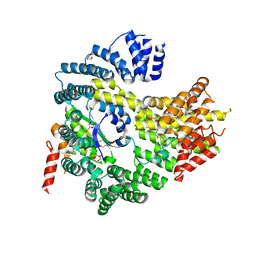 | | Apo N-terminal acetyltransferase complex A | | Descriptor: | GLYCEROL, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1, ... | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
4HNX
 
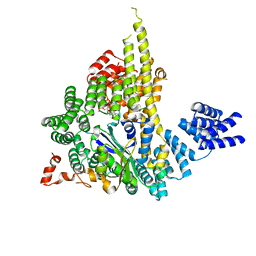 | | The NatA Acetyltransferase Complex Bound To ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1 | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
